Enzymes – Biochemistry – Flashcards
Unlock all answers in this set
Unlock answersquestion
enzymes
answer
important biological catalysts Key features: lower activation energy increase rate of reaction (kinetics) do not alter equilibrium constant are not changed or consumed in the reaction (appear in both reactants and products) --> far fewer enzymes are required relative to overall amount of substrate because one enzyme can act on many molecules of substrate over time are pH- and temperature sensitive, with optimal activity at specific pH ranges and temperatures do not affect overall delta G of reaction are specific for a particular reaction or class of reactions
question
catalysts
answer
do not impact thermodynamics of biological reaction. The deltaH(rxn) and equilibrium position do not change. Help reaction proceed at much faster rate
question
enzyme specificity
answer
enzymes are picky Molecules upon which enzyme act are called substrates. A given enzyme will only catalyze a single reaction or class of reactions with these substrates.
question
Oxidoreductases
answer
catalyze oxidation-reduction reactions. The transfer of electrons between biological molecules. Often have a cofactor that acts as electron carrier (i.e. NAD+ and NADP+) Enzymes with dehydrogenase or reductase - oxidoreductasess. Enzymes in which oxygen is final electron acceptor often include oxidase in their names electron donor - reductant electron acceptor - oxidant
question
reductant
answer
electron donor in reactions catalyzed by oxidoreductases
question
oxidant
answer
electron acceptor in reactions catalyzed by oxidoreductases
question
Transferases
answer
type of enzyme that catalyzes the movmeent of functional group from on emolecule to another. I.e. aminotransferase converts aspartate and alpha ketoglutarate as a pair to glutamate and oxaloacetate. most kinases are a member of this classes.
question
kinases
answer
most are transferases they catalyze transfer of a phosphate group from ATP to another molecule, generally
question
hydrolases
answer
catalyze breaking of a compound into two molecules using addition of water. Many hydrolases are named only for their substrate, i.e. phosphatase: cleaves phosphate group from another moelcule. i.e. peptidases, nucleases, lipases, and phosphatases
question
Lyaes
answer
catalyze cleavage of a single molecule into two products. Do not require water as a substrate and do not act as oxidoreductases. Most enzymes can catalyze reverse of specific reaction so synthesis of two molecules into a single molecule can also be catalyzed by lyase (synthases)
question
Isomerases
answer
catalyze rearrangement of bonds within a molecule. Some isomerases can be classified as oxidoreductases, transferases, or lyases, depending on mechanism of enzyme.
question
Ligases
answer
catalyze addition or synthesis reactions, generally between large similar molecules and often require ATP. Synthesis reactions with smaller molecules accomplished by lyases. encountered in nucleic acid synthesis
question
Mnemonic for Major Enzyme Classifications
answer
LI'L HOT Ligase Isomerase Lyase Hydrolase Oxidoreductase Transferase
question
endergonic reaction
answer
one that requires energy input (delta G ; 0)
question
exergonic reaction
answer
energy is given off (delta G ; 0)
question
activation energy
answer
catalysts make it easier for substrate to reach tarnsition state.
question
substrate
answer
molecule upon which an enzyme acts is known as substrate
question
enzyme-substrate complex
answer
physical interaction between enzyme and substrate is called enzyme substrate complex
question
active site
answer
location within enzyme where substrate is held during chemical reaction. It assumes defined spatil arrangement in the enzyme substrate complex, which dictates specificity of enzyme for molecule or group of molecules. hydrogen bonding, ionic interactions, transient covalent bonds stabilize spatial arrangement --> contribute to enzyme efficiency
question
lock and key theory
answer
suggests enzyme's active site (lock) is already in appropriate confirmation for substrate (key) to bind --> substrate can easily fit into active site like key into lock or hand into glove. No alterations of tertiary or quaternary structure is necessary upon binding of substrate
question
induced fit model
answer
more scientifically accepted than lock key. Substrate has induced change in shape of enzyme. This interaction requires energy and therefore, this part of reaction is endergonic. Letting go of enzyme is easy and doesn't require extra energy so it is exergonic. (transition state
question
cofactors/coenzymes
answer
enzymes require nonprotein molecules called cofactors or coenzymes to be effective. They tend to be smaal so they can bind to active site of enzyme and participate in catalysis of the reaction by carrying charge though ionization, protonation, or deprotonation. usually kept in small concentrations in cell so they can be recruited when needed. attached in many ways: weak noncovalent interactions to strong covalent ones
question
apoenzymes
answer
enzymes without their cofactors
question
holoenzymes
answer
enzymes with cofactors attached
question
prosthetic group
answer
tightly bound cofactors or coenzymes that are necessary for enzyme function
question
cofactors
answer
generally small organic groups, vast majority of them are vitamins or derivatives of vitamins .e. NAD+, FAD, and coenzyme A.
question
water soluble vitamins
answer
B complex vitamins and ascorbic acid (vitamin C) and are important coenzymes that must be replenished regularly because they are easily excreted
question
fat soluble vitamins
answer
A, D, E, and K - regulated by partition coefficients, which quantify ability of molecule to dissolve in a polar s nonpolar environment
question
B1
answer
thiamine
question
B2
answer
riboflavin
question
B3
answer
niacin
question
B5
answer
pantothenic acid
question
B6
answer
pyridoxal phosphate
question
B7
answer
biotin
question
B9
answer
folic acid
question
B12
answer
cyanocobalamin
question
saturation
answer
when enzyme cannot go any faster
question
maximum velocity
answer
at saturation, enzyme is working at maxium velocity (vmax) to increase vmax, increase enzyme concentration. This can be induced by inducing expression of gene encoding the enzye.
question
Michaelis-Menten Plot of Enzyme Kinetics
answer
as amount of substrate increases, enzyme is able to increase its rate of reaction until it reaches maximum enzymatic reaction rate (vmax); once vmax is reached,adding ore substrate will not increase rate of reaction
question
Michaelis Menten Equation
answer
describes how rate of reaction, v, depends on concentration of both enzyme (E) and substrate (S) which forms product (P). When reaction rate is vmax, Km = S. when substrate concentration is less than Km, substrate concentration will greatly affect reaction rate. At high substrate concentration exceeding Km, reaction rate increases much more slowly as it approaches Vm where it becomes independent of substrate concentration normally hyperbola. If sigmoidal (s shaped) due to cooperativity among substrate binding sites
question
Km
answer
substrate concentration at which half the enzyme's active sites are full. It is the Michaelis constant and is often used to compare enzymes. When comparing two enzymes, the one with higher Km has a lower affinity for its substrate because it requires a higher substrate concentration to be half-saturated. Km is an intrinsic property, cannot be altered by changing concentration of substrate or enzyme
question
Lineweaver Burk Plots
answer
double reciprocal graph of Michaelis-Menten equation. Same data graphed in this way yield straight line. intercept with x axis = -1/km intercept with y axis = 1/vmax useful for determining type of inhibition an enzyme is experiencing because vmax and Km can be compared without estimation
question
cooperative enzymes
answer
have multiple sub-units and multiple active sites. Subunits and enzyme may exist in one of two states: low affinity tense state (T) or high affinity relaxed state (R). Binding of substrate encourages transition of other subunits from T state to R state --> increased likelihood of substrate binding other subunits. loss of substrate --> transition from R state to T state and promotes dissociation of substrate from remaining subunits.
question
environmental factors that affect enzyme
answer
temperature acidity alkalinity high salinity
question
enzyme actvity
answer
also known as enzyme velocity/enzyme rate
question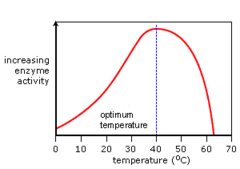
temperature and enzyme actiivty

answer
enzyme catalyzed reactions double in velocity for every 10 degree increase in temperature until optimum temperature reached (37 degrees). After this, activity falls off sharply and enzymes denature. Some enzymes can regain function if cooled after overheating (i.e. Siamese cats, dark on face ears and tails because enzyme responsible for pigmentation is mutated in Siamese cats. Ineffective at body temp but coler temperatures becomes active. Outer extremities are cooled by air --> active form of enzyme and dark)
question
pH and enzyme
answer
optimal pH is 7.4 for humans. Below 7.35 is acidemia.
question
salinity and enzyme
answer
altering concentration of salt can change enzyme activity in vitro.Increasing levels of salt can disrupt hydrogen and ionic bonds --> partial change in conformation of enzyme and in some cases causing denaturation
question
feedback regluation
answer
enzymes are often subject to regulation by products further down a given metabolic pathway.
question
feed-forward regulation
answer
enzymes may be regulated by intermediates that precede enzyme in pathway (much less common than feedback regulation)
question
negative feedback
answer
feedback inhibition, helps maintain homeostasis. Once we have enough product, we turn off pathway rather than creating more. The product may bind to active site of enzyme or multiple enzymes that acted earlier on its biosyntehtic pathway.
question
reversible inhibition
answer
4 types: competitive noncompetitive mixed uncompetitive
question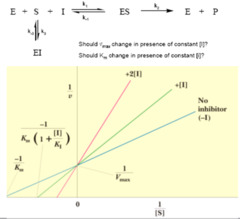
competitive inhibition

answer
occupancy of active site. Substrates cannot access enzymatic binding site if there is an inhibitor in the way. It can be overcome by adding more substrate so substrate to inhibitor ratio is higher. Adding competitive inhibitor, does not alter value of vmax because if enough substrate is added, it will outcompete inhibitor and be able to run reaction to maximum velocity.
question
noncompetitive inhitors
answer
bind to allosteric site instead of active site, which changes enzyme conformation. Two molecules do not compete for same site, so inhibition is considered noncompetitive and cannot be overcome by adding more substrate. Noncompetitive inhibitors bind equally well to enzyme and enzyme-substrate complex, unlike mixed inhibitors. Once enzyme conformation altered, no extra substrate will be helpful. Noncompetitive inhibitor --> decreases vmax because less enzyme available to react. Does not alter Km because any copies of enzyme that are still active maintain same affinity to substrate.
question
mixed inhibition
answer
when inhibitor can bind to either enzyme of enzyme substrate complex but has different affinity for each. If inhibitor has same affinity - noncompetitive inhibitor. Mixed inhibitors do not bind on active site, they bind on allosteric site. It alters Km depending on preference of inhibitor for enzyme vs enzyme-substrate complex. If inhibitor preferentially binds to enzyme, it increases Km value (lowers affinity). If inhibitor binds to enzyme-substrate complex, it lowers Km value (increases affinity) both cases, vmax is decreased.
question
uncompetitive inhibition
answer
binds only to enzyme-substrate complex and locks substrate in enzyme, preventing release. It increases affinity between enzyme and substrate. Because complex has already formed upon binding, uncompetitive inhibitors must bind at an allosteric site. Formation of enzyme substrate complex --> conformation change --> uncompetitive inhibitor can bind. Lowers Km and Vmax
question
irreversible inhibition
answer
active site is unavailable for prlonged periods of time or enzyme is permanently altred i.e. allosteric enzymes, covalently modified enzymes, zymogens
question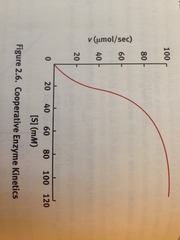
alllosteric enzymes

answer
multiple binding sites on enzyme. Active site is present and allosteric site for regulation of availability alternate between active and inactive form have sigmoidal Michaelis-Menten plot
question
allosteric activator
answer
molecule that binds to allosteric site that causes shift that makes active sites more available
question
allosteric inhibitor
answer
shift that makes active site less available
question
covalently modified enzymes
answer
enzymes can be activated or deactivated by phosphorylation or dephosphorylation
question
glycosylation
answer
covalent attachment of sugar moieties is a covalent enzyme modification. It can tag an enzyme for transport within the cell or can modify protein activity and selectivity
question
zymogens
answer
contain catalytic domain (active) and regulatory domain. This domain must either be removed or altered to expose active site. Apoptotic enzymes exhibit similar regulation. i.e. trypsinogen



