MCAT: Biochemistry – Flashcards
Unlock all answers in this set
Unlock answersquestion
molecules that contain: 1. an amino group (-NH2) 2. carboxyl group (-COOH) 3. R group/side chain - determines the chemical properties 4. Hydrogen atom (-H) main focus is on alpha aa's - not every amino acid is an alpha aa (GABA) - not every amino acid is coded for a codon - some aa's are converted to non codon coded aa for specific reasons - in eukaryotes they are all L configuration
answer
amino acid
question
aa that the amino group and the carboxyl group are attached to the same carbon (alpha carbon of the carboxylic acid) alpha carbon is chiral (beisdes glycine) - optically active - in eukaryotes they are all L configuration
answer
alpha amino acid
question
side group of the amino acid every amino acid has a different one determines the properties of the amino acids and thus their functions
answer
R group
question
gamma aminobutyric acid amino group and carboxyl group are not on the same carbon amino group is on the gamma carbon from the carbonyl
answer
GABA
question
20 alpha amino acids amino acids that are encoded by the human genetic code through codons since they are all chiral at the alpha carbon (besides glycine) - L CONFIGURATION have at least two group that can be protonated/deprotonated - two (maybe three) pka's
answer
proteinogenic amino acids
question
the alpha carbon in alpha aa's optically active has four different groups attached to it exception - glycine (has two H's) L configuration - goes to the S absolute configuration
answer
chiral amino acids
question
Glycine *GLY* R group - H ACHIRAL (not L or D) smallest amino acid *nonpolar, non-aromatic R group* hydrophobic - interior of proteins
answer
G
question
L amino acids are S configuration AMINO GROUP IS TO THE LEFT in a fisher projection
answer
Cahn Ingold Prelog System
question
Cysteine *CYS* R group = -CH2SH *polar side chain* has a *thiol* group -since sulfur is larger and less electronegative than oxygen SH bond is weaker than an OH bon -makes SH prone to OXIDATION reaction L amino acid *R absolute configuration *
answer
C
question
Alanine *ALA* R Group = -CH3 *nonpolar, non-aromatic R group* alkyl side chain with one carbon makes it nonpolar L amino acid S absolute Configuration hydrophobic - interior of proteins
answer
A
question
Arginine *ARG* R group = -CH2CH2CH2-NH-C(=NH)NH2 *positive/basic Side Chain* -three nitrogens in its side chain *positive charge is delocalized over all three nitrogen atoms* L amino acid S absolute Configuration hydrophilic - surface of proteins
answer
R
question
Asparagine *ASN* R group = -CH2C(O)NH2 *polar side chain* *amide side chain* L amino acid S absolute Configuration hydrophilic - surface of proteins
answer
N
question
Aspartic Acid/ ASPARTATE *ASP* R group = -CH2C(O)OH *Negative/Acidic Side Chain* L amino acid S absolute Configuration hydrophilic - surface of proteins
answer
D
question
Glutamic Acid/ GLUTAMATE *GLU* R group = -CH2CH2C(O)OH *Negative/Acidic Side Chain* L amino acid S absolute Configuration hydrophilic - surface of proteins

answer
E
question
Glutamine *GLN* R group = -CH2CH2C(O)NH2 *polar side chain* *amide side chain* L amino acid S absolute Configuration hydrophilic - surface of proteins
answer
Q
question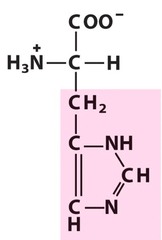
Histidine *HIS* R group = imidazole *positive/basic Side Chain* -aromatic ring with two nitrogens L amino acid S absolute Configuration hydrophilic - surface of proteins

answer
H
question
Isoleucine *ILE* R group = -CH(CH3)CH2CH3 *nonpolar, non-aromatic R group* terminal primary amino group L amino acid alkyl side chain with four carbons makes it nonpolar S absolute Configuration hydrophobic - interior of proteins
answer
I
question
Leucine *LEU* R group = -CH2CH(CH3)CH3 *nonpolar, non-aromatic R group* alkyl side chain with four carbons makes it nonpolar L amino acid S absolute Configuration hydrophobic - interior of proteins
answer
L
question
Lysine *LYS* R group = -CH2CH2CH2CH2NH2 *positive/basic Side Chain* L amino acid S absolute Configuration hydrophilic - surface of proteins
answer
K
question
Methionine *MET* R group = -CH2CH2SCH3 *nonpolar, non-aromatic R group* - contains a sulfur group - nonpolar because methyl group is attached to sulfur L amino acid S absolute Configuration
answer
M
question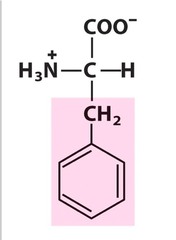
Phenylalanine *PHE* R group = -benzyl *aromatic R group* relatively *nonpolar* L amino acid S absolute Configuration hydrophobic - interior of proteins

answer
F
question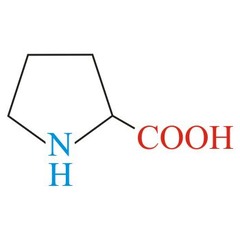
Proline *PRO* R group = five membered ring (amino nitrogen becomes apart of the side chain) *nonpolar, non-aromatic R group* -restricts the flexibility of it -will form a kink in the secondary structure when found in the middle of an alpha helix - rarely found in alpha helixes (besides the ones that cross the cell membrane) -rarely found in beta pleated sheets - only found in the turns of the beta pleated sheets -found at the start residue of the alpha helix L amino acid S absolute Configuration

answer
P
question
Serine *SER* R group = -CH2OH *polar side chain* contains an OH group - makes them highly polar and able to participate in hydrogen bonding L amino acid S absolute Configuration
answer
S
question
Threonine *THR* R group = -CH(OH)CH3 *polar side chain* contains an OH group - makes them highly polar and able to participate in hydrogen bonding L amino acid S absolute Configuration
answer
T
question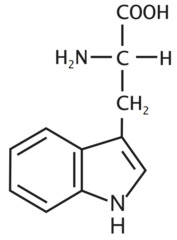
Tryptophan *THR* R group = five ring(with nitrogen) - benzene ring *aromatic R group* -double ring system with nitrogen L amino acid S absolute Configuration

answer
W
question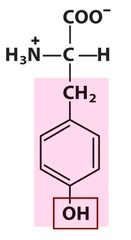
Tyrosine *TRY* R group = benzyl -OH *aromatic R group* -relatively *polar* due to the OH but AROMATIC L amino acid S absolute Configuration

answer
Y
question
Valine *VAL* R group = -CH(CH3)CH3 *nonpolar, non-aromatic R group* alkyl side chain with three carbons makes it nonpolar L amino acid S absolute Configuration hydrophobic - interior of proteins
answer
V
question
Glycine Alanine Valine Leucine Isoleucine Methionine Proline alkyl side chain with one, two, three or four carbons

answer
Nonpolar, Non-aromatic Amino Acids
question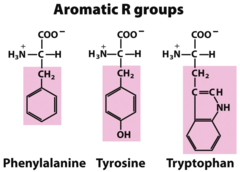
tryptophan phenylalanine tyrosine

answer
Aromatic Amino Acids
question
CH2 + benzene ring
answer
benzyl
question
serine threonine asparagine glutamine cysteine POLAR but NOT aromatic OH AMIDE SIDE GROUP THIOL
answer
Polar side chain amino acids
question
C(O)NH2 in asparagine and glutamine the nitrogens DO NOT gain/lose protons with a change in pH they DO NOT become charged
answer
amide side chains
question
-SH cysteine since S is larger and less electronegative than Oxygen, SH bonds are weaker than OH bonds (why cysteine is prone to oxidation reactions)
answer
Thiol
question
Aspartic acid glutamic acid GLUTAMATE - ion/deprotonated version of glutamic acid ASPARTATE - ion/deprotonated version of aspartic acid - most acids in cells exist in the deprotonated form
answer
Negatively Charged/ Acidic Amino Acids
question
arginine lysine histidine all side chains has positively charged nitrogen atoms
answer
Positively Charged/ Basic Amino Acids
question
side chain of histidine aromatic ring with two nitrogens pka of side chain is 6 - so at a physiological pH the side chain is neutral amide then will be protonated at a higher pH
answer
imidazole
question
found in the interior proteins alanine leucine valine phenylalanine when places in a aqueous solution - water molecules in the solvation layer cannot form hydrogen bonds with the side groups this makes water molecules have to rearrange themselves to maximize hydrogen bonding - this is them increasing order - thus *decreasing in entropy/ chaos) - makes the overall process non spontaneous (+G)
answer
hydrophobic amino acids
question
surface of proteins charged or polar amino acids histidine lysine serine arginine glutamic acid aspartic acid asparagine glutamine putting these residues on the exterior of protein allows water molecules more freedom in their positioning increasing entropy (S) (increasing chaos) making the overall solvation process spontaneous (-G)
answer
hydrophilic amino acids
question
can either accept or donate a proton how they react depends on their enviroment amino acids are these because of their acidic carboxyl group and basic amino group 1. ionizable groups tend to gain protons when in LOW pH/ ACIDIC conditions = *protonated* 2. ionizable groups tend to lose protons when in HIGH pH/BASIC conditions = *deprotonated* 3. pka; pH (pH is LOW) - protonated 4. pka; pH (ph is HIGH) - deprotonated
answer
amphoteric species
question
pH where half of the molecules of that species are deprotonated protonated version = deprotonated version HA = A-
answer
pka
question
carboxyl group usually around two pka;pH (acidic/ LOW pH/ lots of H) COOH pka;pH (basic/ HIGH pH/ few of H) COO-
answer
pka1
question
amino group usually around nine pka;pH (acidic/ LOW pH/ lots of H) NH3+ pka;pH (basic/ HIGH pH/ few of H) NH2
answer
pka2
question
R group pka greater than
answer
pka3
question
pka;pH (acidic/ LOW pH/ lots of H) COOH pka;pH (acidic/ LOW pH/ lots of H) NH3+ POSITIVE
answer
amino acid structure in acidic pH
question
pka;pH (basic/ HIGH pH/ few of H) COO- pka;pH (basic/ HIGH pH/ few of H) NH2 NEGATIVE
answer
amino acid structure in basic pH
question
pH is in between the pka's of the carboxyl group and the amino group has both a negative charge and a positive charge pka pH of the amino group the two charges neutralize each other exist in water as internal salts
answer
zwitterions
question
amino acids are great candidates for titrations because they have both acid and base properties two curves if no charged side chain three curves if have charged side chain low pH = fully protonates (POSITIVE CHARGE) = NH3+, COOH middle pH = zwitterion (NEGATIVE + POSITIVE CHARGE) = NH3+, COO- high pH = fully deprotonated (NEGATIVE CHARGE) = NH2, COO- pH = pka1 when COOH --> COO- SOLUTION IS ACTING AS A BUFFER horizontal graph pH = pI MOLECULE IS SENSITIVE TO PH CHANGES vertical graph pH= pka2 when NH3+--> NH2 SOLUTION IS ACTING AS A BUFFER horizontal graph
answer
titrations of amino acids
question
isoelectric point the pH at which the molecule is electrically neutral! molecule is sensitive to pH changes vertical part on the titration curve for no charged side chain equation is..
answer
pI
question
aspartate and glutamate LOW pH fully protonated state = +1 first Deprotonation is on carboxyl group --> now neutral send Deprotonation is on the R group --> now -1 third deprotonation is on the amino group is much LOWER than the pI of an amino acid with no charged group
answer
pI acidic amino acid
question
arginine, lysine, histidine LOW pH fully protonated state = +2 first Deprotonation is on carboxyl group --> now +1 send Deprotonation is on main amino group --> now -1 third deprotonation is on the amino group of the side chain is much HIGHER than the pI of an amino acid with no charged group
answer
pI basic amino acid
question
composed of amino acid residues
answer
peptides
question
amino acid subunits make up peptides
answer
residues
question
two aa residues
answer
dipeptides
question
three aa residues
answer
tripeptides
question
small peptides with no more than 20 aa residues
answer
oligopeptides
question
peptides that have more than 20 aa residues
answer
polypeptides
question
joins together amino acid residues specialized form of an amide bond forms between the COO- group of the second amino acid and the NH3+ of the first amino acid neutrophilic nitrogen of NH3+ attacks the electrophilic carbonyl carbon of COO- hydroxyl group of the carboxylic acid is kicked off - water is a end product -C(O)NH- functional group because the amide group have delocalized pi electrons in the carbonyl and the lone pair on the amino nitrogen they can show resonance - C-N bond in the amide has a partial double bond character -C-N bond is more restricted - making the protein more rigid - rotation can happen around the other bonds because they are sigma bonds formation = * condensation or dehydration reaction* - removal of water *acyl substitution reaction*
answer
peptide bonds
question
on the first amino acid it is the free amino terminus
answer
N terminus
question
on the last amino acid in the chain free carboxyl end is this
answer
C terminus
question
cleaves at the carboxyl end of arginine and lysine break it apart by adding a hydrogen to the amide nitrogen and Oh group to the carbonyl carbon
answer
trypsin
question
cleaves at the carboxyl end of the phenylalanine, tryptophan, and tyrosin break it apart by adding a hydrogen to the amide nitrogen and Oh group to the carbonyl carbon
answer
chymotrypsin
question
linear arrangement of amino acids which is coded by DNA sequence of amino acids from the N terminus to the C terminus stabilized by the formation of *covalent peptide bonds* between the adjacent amino acids encodes for all the information needed for folding at all the higher structural levels - all other levels are more energetically favorable arrangements
answer
primary structure of peptides
question
local structure of neighboring amino acids is a result of *hydrogen bonds* between the near by amino acids stability is due to the formation of *intramolecular hydrogen bonds* alpha helices Beta pleated sheets
answer
Secondary structure of peptides
question
form of a secondary structure of peptides rod like structure peptide chain coils clockwise around the central axis is stabilized by the carbonyl oxygen atom and an amide hydrogen atom four residues down -- hydrogen bonding side chains of the amino acids point away from the helical core ex. keratin
answer
alpha helices
question
form of a secondary structure of peptides can be parallel or anti-parallel peptide chains that form rows or strands held together by intramolecular hydrogen bonds between the *carbonyl oxygen* one of chain and the (amide hydrogen* on the adjacent chain to accommodate as many hydrogen bonds they have a rippled shape the R groups point below or above the sheet
answer
beta pleated sheets
question
ex. collagen have structures that resemble sheets or long strands
answer
fibrous proteins
question
ex. myoglobin tend to be spherical
answer
globular proteins
question
creates the tertiary and quaternary protein structure s
answer
protein folding
question
three dimensional shape determined by 1. *hydrophobic/ hydrophilic* interactions between *R group*s of amino acids ----hydrophobic residues - interior of proteins (A, I, L, V, F) - reduces their proximity to water ----hydrophilic residues - surface of proteins (E, D, N, Q, H, K, R) - ----------hydrophilic NH and C=O bonds found in the polypeptide chain get pulled in by the hydrophobic residues ---------hydrophilic bonds can then form electrostatic interactions and hydrogen bonds that further stabilize the protein from the inside *hydrophobic and hydrophilic interactions cause it to collapse into the proper arrangement* 2. hydrogen bonding 3. Acid and Base Interactions between R groups (creating salt bridges) or disulfide bonds
answer
tertiary structure of peptides
question
bond that forms when two cysteine molecules become oxidized to form cystine create loops in the protein chain (determine how curly hair is) requires the loss of two protons and two electrons oxidation reaction
answer
disulfide bonds
question
oxidized form of two cysteines has a disulfide bond in it
answer
cystine
question
intermediate state in between the secondary structure and the tertiary structure
answer
molten globules
question
protein loses its tertiary structure loses its function often irreversible unfolded proteins cannot catalyze reactions causes: 1. heat - temperature increases, average kinetic energy increases (increased molecular motion) -extra energy can be enough to overcome the hydrophobic interactions that hold the protein together (causing it to unfold) 2. solutes -directly interfere with the forces that hold the protein together - can disrupt the tertiary and quaternary structures by breaking the disulfide bridges (reducing cystine back to cysteine) -can overcome hydrogen bonds and other interactions that hold the alpha helices and beta pleated sheets together
answer
denaturation
question
layer of solvent molecules that surround the solute when a solute is dissolved in a solvent hydrocarbons are more stable in water than organic ones making enthalpy negative (-H)
answer
solvation layer
question
not all proteins have this type of structure only exist for proteins that contain more than one polypeptide chain the arrangement of smaller globular peptides/subunits this is the functional form of the protein ex. hemoglobin and immunoglobulins functions: 1. can make it more stable by reducing the surface area of the protein complex 2. reduce the amount of DNA needed to encode the protein complex (viruses minimize their DNA because on protein is repeated hundreds of times) 3. brings catalytic sites close together - allowing intermediates from one reaction to be directly shuttled to a second reaction 4. can induce *cooperativity/ allosteric effects* - undergo a conformational change to enhance/reduce the activity of the other subunits
answer
Quaternary structure
question
part of their function comes from the prosthetic groups which are covalently attached to them
answer
conjugated proteins
question
cofactors that are strongly covalently attached to the enzyme necessary for the enzyme to function properly can be: organic molecules (vitamins, metal ions) lipids carbohydrates nucleic acids have major roles in determining the function of their respective proteins ex. hemoglobins prosthetic group is HEME - contains iron at the core so oxygen can bind to it can also make the protein it is attached to go to a specific location (cell membrane, nucleus, lysosome, ER)
answer
prosthetic groups
question
proteins with carbohydrate prosthetic groups
answer
glycoproteins
question
solubilize proteins results in a hydrophobic core that promotes denaturation of the protein
answer
SDS
question
biological catalysts DO NOT impact the thermodynamics of the reaction lower the activation energy increase the rate of the reaction do not alter the equilibrium constant are not changed or consumed in the reaction (will appear as a reactant and a product) are pH and temperature sensitive - optimal activity is at a specific pH and temperature do not effect the overall ?G are specific for a particular reaction or class of reactions (*enzyme specificity*) can effect how quickly the reaction gets to equilibrium but not the actual equilibrium state itself Only effects the KINETICS (rate) lower the ACTIVATION ENERGY - make it easier for the substrate to react the transition state Types: 1. oxidoreducatses 2. transferases 3. hydrolases 4. Lyases 5. isomerases 6. Ligases may act to provide a *favorable microenviroment* through *charge or pH* *stabilize the transition state* *bring reactive groups near to one another in the active site*
answer
enzymes
question
molecules that enzymes act upon
answer
substrates
question
enzyme that catalyzes oxidation reduction reactions transfer of electrons between biological molecules often have a cofactor that acts as a electron carrier (NAD+ and NADP+) contains reductant and oxidant enzymes with *dehydrogenase* or *reductase* are these type of enzymes *oxidases* are also these type of enzymes and oxygen is the final electron acceptor
answer
oxidoreducatses
question
electron donor
answer
reductant
question
electron acceptor
answer
oxidant
question
type of enzymes that catalyze the movement of a functional group from one molecule to another ex. *kinases*
answer
transferase
question
under the transferase type of enzyme transfer the phosphate group (usually from ATP) to another molecule
answer
kinase
question
type of enzymes that catalyze the breaking of a compound into two molecules using the addition of water usually named after their substrate ex. phosphatase, peptidases, nucleases, lipases break down proteins, nucleic acids and lipids
answer
hydrolases
question
a hydrolase cleaves a phosphate group from another molecule
answer
phosphatase
question
type of enzymes that catalyze the cleavage of a single molecule into two products NO NOT REQUIRE WATER DO NOT ACT AS OXIDOREDUCTASES can make two molecules into one molecule by LYASE - when doing this they are known as *synthases* usually form double bonds or cyclic molecules to counter act the fact that it does not transfer electrons or use water
answer
lyases
question
type of enzyme that catalyze the rearrangement of bonds within a molecule can catalyze reactions between sterioisomers and constitutional isomers depending on the mechanism - they can also be classified into oxidoreductases, transferases, or lyases
answer
isomerases
question
type of enzyme that catalyze the addition or synthesis reactions - usually between larger and similar molecules usually need ATP (lyases are with smaller molecules) usually seen in nucleic acid synthesis and repair
answer
ligases
question
reaction that REQUIRES energy input +?G reactants lower in energy than products
answer
endergonic
question
reaction which energy is GIVEN OFF -?G reactants higher in energy than products
answer
exergonic
question
-?G
answer
spontaneous reaction
question
physical interaction between the substrate and the enzyme
answer
enzyme substrate complex
question
location within the enzyme where the substrate is held during the chemical reaction assumes a defined spatial arrangement in the enzyme substrate complex dictates the specificity of that enzyme for a molecule/ group of molecules hydrogen bonding/ ionic interactions/ covalent bonds in this stabilize the spatial arrangement and contribute to the efficiency of the enzyme
answer
active site
question
theory for enzyme substrate binding less accepted theory active site is the lock and the substrate is the key the substrate can easily fit into the active site there is no alteration of the tertiary or quaternary structure which is necessary upon binding of the substrates NO CONFORMATIONAL CHANGE perfect fit
answer
Key and Lock Theory
question
theory for enzyme substrate binding more accepted theory both the substrate and the active site change conformation the substrate induces a change in shape of the enzymes the initial change requires energy (part of the reaction is endergonic) the product is more relaxed version and does not require any reaction to leave (exergonic part of the reaction) enzymes go back to their original shape once they are "let go of" first they don't seem like they fit together find that the induced form/ transition state is more comfortable for both of them shape of the active site becomes truly complementary only after the substrate begins to bind with the enzyme substrate of the wrong type will not cause the appropriate conformational shift in the systems active site will not be perfectly shown and the transition site will not be preferred
answer
Induced Fit Modle
question
coenzymes non protein molecules that make the enzyme effective small in size help the substrate better fit to the enzyme can bind to the active site of the enzyme and participate in the catalysis of the reaction usually *carry charge* through ionization, protonation, deprotonation usually kept at low concentrations - recruited only when needed can be attached via weak covalent bonds or by strong covalent bonds
answer
cofactors
question
enzymes without their cofactors
answer
apoenzymes
question
enzymes with their corresponding cofactors
answer
holoenzymes
question
B1
answer
thiamine
question
B2
answer
Riboflavin
question
B3
answer
niacin
question
B5
answer
pantotheic acid
question
B6
answer
pyridoxal phosphate
question
B7
answer
biotin
question
B9
answer
folic acid
question
B12
answer
cyanocobalamin
question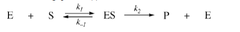
depends on: environmental conditions concentration of enzyme concentration of substrate cannot go any faster when it has hit saturation k1 = rate at which the enzyme substrate complex forms k2 = rate at which ES complex comes apart k3 = rate at which ES forms enzyme and separate product
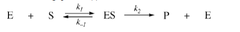
answer
enzyme kinetics
question
when no matter how much substrate you add the enzyme is working at the maximum velocity (Vmax) only way to increase the Vmax at this point is to increase the enzyme concentration
answer
enzyme saturation
question
V = rate of reaction km = michaelis constant
answer
Michaelis menten equation
question
equals the solute concentration when at 1/2 vmax when 1/2Vmax Km= [S] Michaelis constant often used to compare enzymes *measure of the affinity of the enzyme for its substrate* substrate solution when HALF OF ALL THE ENZYMES CONTAIN SUBSTRATE higher Km = lower affinity because you need more substrate to bind achieve half of the vmax it is an instrinsic property of the enzyme substarte complex cannot be altered by changing the concentartion of the susbtarte or the enzyme
answer
Km
question
for a given concentration of an enzyme - it usually is a hyperbola when substrate concentration is less the km = changes in substrate concentration will GREATLY affect the reaction rate when substrate concentration is more than km = the rate of the reaction increases much more slowly as it approaches vmax - until it becomes independent of substrate concentration
answer
Michaelis menten relationship
question
double reciprocal graph of the Michaelis Menten equation yields a straight line x axis = -1/km *decreased if moves left* y axis = 1/vmax *decreased if moved up* useful when determining the type of INHIBITION that an enzyme is experiencing
answer
Lineweaver Burk Plots
question
when graphed in the michaelis menten graph they do NOT show the hyperbola but and SIGMOIDAL (s shaped) curve these enzymes have multiple binding subunits and multiple active sites the subunits could be in the T state or the R state binding of the substrate encourages the transition of the other subunits to go from the T state to the R state - this will increase the likelihood of the substrate binding by these other subunits - loss of the substrate can encourage the R state to go into the T state
answer
cooperativity
question
tense state of a enzyme subunit low affinity state
answer
T state
question
relaxes state of an enzyme subunit high affinity state
answer
R state
question
enzyme velocity = enzyme rate rate when all enzymes are occupies - has to do with efficiency greater is a greater efficiency
answer
vmax
question
double in velocity for ever 10 degrees Celsius until optimal temperature is reached human body optimal temp is 37C/98.6F/310K activity falls sharply after that because proteins start to become denatured siamese cats - tyrosinase (enzyme responsible for pigmentation) is mutated - active at cooler temps and inactive at body temp pigmentation only in the cooler lower extremities
answer
enzyme activity and temperature
question
pH effect the ionization of the active site changes in it can lead to denaturation optimal active at 7.4 exceptions stomach pepsin - ph = 2 pancreatic enzymes pH= 8.5
answer
enzyme activity and pH
question
salt concentration can change the enzyme activity in vitro increasing concentration can disrupt hydrogen and ionic bonds this can cause partial changes in the conformation of the enzyme also can lead to denaturation
answer
enzyme activity and salinity
question
regulation by products further down/or precede a given metabolic pathway precede - feed forward further down - negative feedback - can bind to the active site or to other enzymes that acted earlier many hormonal feedback loops demonstrate negative feedback
answer
feedback regulation
question
four types competitive non competitive mixed uncompetitive
answer
reversible inhibition
question
the inhibitor takes up the *active site space* substrates cannot access the enzymatic binding sires if there is inhibitor in the way to over come this you have to add more substrate thus the substrate to inhibitor ratio must be higher if more molecules of substrate are available than molecules of inhibitor then the enzyme will be more likely to bind substrate than the inhibitor *Vmax is UNCHANGED* - stays - if enough substrate is added, it will outcompete the inhibitor and be able to run the reaction at maximum velocity *Km is INCREASED* - goes right - substrate concentration has to be higher to reach half of the maximum velocity in the presence of the inhibitor
answer
Competitive Inhibition
question
binds to an *allosteric site* this indices a change in the enzymes conformation substrate and the inhibitor do NOT compete for the same space bind equally well to the enzyme as to the enzyme substrate complex *same affinity for both sites* once the enzyme's conformation has changes - no extra amount of substrate will be good enough to make the enzyme substrate complex *Vmax is DECREASED* - goes up - there is less enzyme available to react* *Km is UNCHANGED* - stays - any copes of the enzyme that are still active will have the same affinity for their substrate
answer
Non Competitive Inhibition
question
inhibitor can bind to either the enzyme or the enzyme substrate complex but has a different affinity for the enzyme and for the ES complex bind an *allosteric site* *Km is ALTERED* depends on the preference of the inhibitor to the E vs ES *higher affinity for E = Km will INCREASE (lowers affinity)* *higher affinity for ES = Km will DECREASE (increase affinity)* - will not let the substrate go *Vmax is always DECREASED* not parallel - doesn't converge on an axis
answer
Mixed Inhibition
question
inhibitors only binds to the *ES complex* essentially locks the substrate in the enzyme - and prevents it's release increasing the affinity between the substrate and the enzyme because the ES complex has already been formed they bind to *allosteric sites* actually the binding of the substrate creates a conformational change that allows the uncompetitive inhibitor to bind *Km and Vmax is DECREASED* will be in parallel
answer
Uncompetitive Inhibition
question
non catalytic regions of the enzyme that binds regulators
answer
allosteric sites
question
in this the active site is unavailable for a prolonged period of time or the enzyme is permanently altered this type of inhibition is not easily overcome or reversed ex. asprin prime drug mechanism new enzymes must be transcribed and translated
answer
irreversible inhibition
question
they have multiple binding sites the active site is available as well as at least one other site that can regulate the availability of the active site can alternate between the active and inactive form inactive form - cannot carry out enzymatic activities molecules that bind can be allosteric activators or allosteric inhibitors - binding of either of them will cause a conformational change can alter the activity of the enzyme usually have a sigmoidal shaped S curve
answer
allosteric enzymes
question
enzymes are subject to covalent modifications can be activated or deactivated via phosphorylation or dephosphorylation they can also be glycosylated (attachment of sugar residues)
answer
covalently modified enzymes
question
binds to allosteric enzymes causing a conformational change binding of this will result in a shift that makes the active site form available for binding to the substrate
answer
allosteric activators
question
binds to allosteric enzymes causing a conformational change binding of this will result in a shift that makes the active site LESS AVAILABLE for binding to the substrate
answer
allosteric inhibitors
question
addition of sugar residues on enzymes can tag an enzyme for transport can also modify protein activity or selectivity
answer
glycosylation
question
inactive enzymes enzymes that need to be tightly controlled contain catalytic/active domain and regulatory domain regulatory domain must be removed/altered to expose the active site ex. trypsinogen which is inactive form of trypsin *ogen* suffix
answer
zymogens
question
apoptotic enzymes regulatory and active domain regulatory domain must be removed/ altered to expose the active site similar mechanism to the way zymogens work
answer
caspases
question
three dimensional web/ scaffolding system for the cell proteins are anchored to the cell membrane by the the embedded protein complexes also support the tissues for the body proteins make up the tendons, ligaments cartilage, and basement membranes
answer
cytoskeleton
question
collagen elastin keratin actin tubulin have high repetitive secondary structure highly repetitive motif
answer
primary structural proteins
question
primary structural protein trihelical fiber (three left handed helices woven together to form a secondary right handed helix) makes up the extracellular matrix of connective tissue found throughout the body gives strength and flexibility
answer
collagen
question
primary structural protein component in extracellular matrix in connective tissue stretches and then recoils like a spring restores the original shape of the tissue
answer
elastin
question
primary structural protein intermediate filaments found in epithelial cells contribute to the mechanical integrity functions as a regulatory protein makes up hair and nails
answer
keratin
question
primary structural protein makes up microfilamnets and the think filaments in myofibrils most abundant proteins in eukaryotic cells these proteins have a positive and negative side - polarity allows proteins to travel unidirectionally along the actin filament (one way street)
answer
actin
question
primary structural protein protein that makes up microtubules provide structure chromosome separation in meiosis and mitosis intracellular transport with kinesin and dynein also has polarity (+ and - side) negative side is located adjacent to the nucleus positive side is by the periphery of the cell
answer
tubulin
question
super-secondary structure a repetitive organization of secondary structural elements
answer
motif
question
display enzymatic activity acting as ATPases - they power a conformational change necessary for motor function they have transient interactions with either *actin* or *microtubules* ex. myosin, kinesins, dyeins
answer
motor proteins
question
primary motor protein that interacts with actin thick filament in a myofibril can be involved in cellular transport the single subunit has a neck and head - movement of the neck can create the power stroke in the sarcomere contraction
answer
myosin
question
motor protein associated with microtubules two heads - one must stay attached to tubulin at all times step like walking aligns chromosomes during metaphase and depolymerizing microtubules during anaphase of mitosis important in vesicle transport *bring vesicles toward the positive end of the microtubule (periphery)* bring vesicles of neurotransmitter to the positive end of the axonal microtubule - toward the synaptic terminal
answer
kinesins
question
motor protein associated with microtubules two heads - one must stay attached to tubulin at all times step like walking involved in the sliding movement of cilia and flagella important in vesicle transport *bring the vesicles toward the negative end of the microtubule (nucleus)* bring vesicles of waste or recycled neurotransmitter back toward the end of the microtubule (toward to the soma) through *retrograde transport*
answer
dyeins
question
have stabilizing effects on individual cells and the body they transport or collect molecules through binding to them ex. hemoglobin, calcium binding proteins, DNA binding proteins (usually transcription factors) each of these proteins has an affinity curve for the molecule of interest hemoglobin has a oxyhemoglobin curve curve differs depending on the gaol of the binding proteins sequestration (taking something) of the molecule is the goal - the binding protein usually has high affinity for its target across a large range of concentrations so t can keep it bound at nearly 100% go is to transport the molecule - must be able to bind and unbind to target to maintain steady state concentrations - varying affinity depending on the environmental conditions
answer
binding proteins
question
Cell Adhesion Molecules proteins found on the surface of most cells aid in binding the cell to the extracellular matrix or other cells *all are integral membrane proteins (permanently attached to the membrane) major families: 1. Cadherins 2. Integrins 3. Selectins to stop clotting processes during heard attacks - other drugs target these
answer
CAMs
question
family of CAMs group of glycoproteins that mediate calcium dependent cell adhesion hold similar cell types together (epithelial cells) different cells will have type specific these epithelial cell uses E cadherin nerve cells use N cadherin
answer
Cadherins
question
family of CAMs have a alpha and beta chain that spans the membrane - chains are important because they bind to and communicate with the extracellular matrix play an important role in cellular signaling can impact cellular function by promoting cell division and apoptosis ex. ?IIb ?3 integrin allows the platelets to stick to the fibrinogen - clotting factor. this causes activation of platelets to stabilize the clot others are used for: white blood cell migration stabilization of epithelium on the basement membrane
answer
Integrins
question
family of CAMs bind to carbohydrate molecules that project from other cell surfaces expressed on white blood cells, endothelial cells that line the blood vessels play a huge role in host defense/ inflammation and white blood cell migration
answer
Selectins
question
neutralize the antigen - making the pathogen or toxin unable to exert its effect on the body marking the pathogen for destruction by other while blood cells immediately = opsonization clumping together/ agglutinating the antigen and antibody into large insoluble protein complexes that can be phagocytized and digested by macrophages
answer
anitbody - antigen consequences
question
marking the pathogen for destruction by other while blood cells immediately
answer
opsonization
question
process in which cell receive and act on signals proteins participate in this - acts on extracellular ligands, acts as transporters for facilitated diffusion, receptor proteins and second messengers can take advantage of another existing gradients (ion channels) or second messenger cascades (enzyme linked receptors and G protein coupled receptors)
answer
biosignaling
question
proteins that create specific pathways for charged molecules three typesL 1. Ungated channels 2. Voltage gated channels 3. Ligand Gated Channels Km and Vmax are also applicable here Km - refers to the solute concentration at which the transporter is functioning at half the maximum activity
answer
ion channels
question
type of ion channel have no gates and are unregulated all cells possess ungated potassium channels - there will be a net efflux of potassium through these channels unless potassium is at equilibrium
answer
ungated channels
question
type of ion channel gate is regulated by the membrane potential change near the channel many excitable (neurons) exhibit voltage gated sodium channels - closed under resting conditions but membrane depolarization comes and the protein conformation changes allows the protein channel to open and then quickly close and the voltage increase s ex. by the sinoatrial node of the heart - serve as a pacemaker current also the voltage drops - these channels open to bring the cell back to threshold and the fire another action potential
answer
voltage gated channels
question
type of ion channel binds a specific substance or ligand to the channel causes it to open or close
answer
ligan gated channels
question
membrane receptors that display catalytic activity in response to ligand binding can be auto-reactive have three domains: membrane spanning domain - anchors receptor in the cell membrane ligand binding domain - stimulated by a proper ligand and created a change in the catalytic domain catalytic domain change of the conformation of the catalytic domain usually initiates the second messenger cascade ex. RTK, serine/threoinine specific protein kinases, receptor tyrosin phosphatases
answer
enzyme linked receptors
question
receptor tyrosin kinases example of enzyme linked receptors composed of a monomer that dimeizes upon ligand binding dimer is active form that phosphates additional cellular enzymes - including itself
answer
RTK
question
G protein coupled receptors large family of integral membrane proteins involved in signal transduction trimer dissociation upon activation two protein complex have seven membrane-spanning alpha helices they differ at the specificity of the ligand binding area found in the extracellular surface of the cell they utilize the *heterotrimeric G Protein* names for their intracellular link to guanine nucleotides (GDP and GTP) the binding of the ligand increases the affinity of the receptor for the G protein binding of the G protein represents a switch to the active state and affects the intracellular signaling pathway three types of G proteins : 1. Gs 2. Gi 3. Gq
answer
GPCR
question
type of G protein stimulates *adenylate cyclase* INCREASES the level of cAMP in the cell
answer
Gs
question
type of G protein inhibits *adenylate cyclase* DECREASES the level of cAMP in the cell
answer
Gi
question
type of G protein activates *phospholipase C* - cleaves a phospholipid from the membrane to form PIP2. PIP2 is then cleaved into DAG and IP3. IP3 can open calcium channels in the ER - increases Calcium levels in the cell
answer
Gq
question
the binding of the ligand on the GPCR increases the affinity of the receptor for the G protein binding of the G protein represents a switch to the active state and affects the intracellular signaling pathway three types of G proteins : 1. Gs 2. Gi 3. Gq they have three subunits (alpha beta and gamma) bind to GDP and is in complex with beta and gamma subunits when INACTIVE when ligand bind the GPCR - receptor becomes activated and GTP is formed the alpha subunit becomes dissociated from the beta and gamma subunits activated alpha subunit alters the activity of adenylate cyclase (alpha*s* = activated) once GTP is activated by alpha subunit - will be dephosphorlated to become GDP and the alpha subunit will rebind to the beta and gamma subunits making the G protein inactive
answer
G protein
question
crushing grinding blending
answer
homogenization
question
homogeniztion then centrifugation then isolation electrophoresis and chromatography - both can be used for real or denatured proteins
answer
protein isolation
question
method of separating proteins subjects proteins to an electric field moves the proteins to their *net charge and size* - compounds = move to the anode (+) + compound = move to the cathode (-) uses a electrolytic cell (????G>O and Ecell <O) slower neutral bigger placed in a small electric field faster smaller more charged placed in a bigger electric field
answer
electrophoresis
question
velocity at which the protein will move to the anode (+) or move to the cathode (-) directly proportional to the electric fields strength (E) and the net charge of the molecule (z) and inversely proportional to the frictional coefficient (f)
answer
migration velocity
question
f depends on the mass and shape of the migration molecule in the migration velocity of electrophoresis equation
answer
frictional coefficient
question
standard medium for protein electrophoresis slightly pores mixture solidifies at room temperature molecules will both this at different speeds due to their charge and their size acts as a seive - smaller molecules can go more quickly
answer
polyacrylamide gel
question
negative side of the gel cations always move to it
answer
cathode
question
positive side of the gel anions always move to it
answer
anode
question
polacrylamide gel electrophoresis analyzes proteins in their *native states* limited by the mass to charge and mass to size ratios many different proteins can have the same level of migration native protein can be taken back but only if it is not stained most useful to compare the molecular size or charge of proteins known to be similar in size from other analytic methods like SDS-PAGE o size exclusion chromatography
answer
native PAGE
question
electrophoresis that separates the proteins on mass alone is PAGE but adds SDS - a detergent that disrupts all non covalent interactions creates a large chains with negative charges (everything is -) this neutralizes the proteins original charge and denatures the protein the only thing effecting the velocity - *electric field strength and the frictional coefficient (depends on mass) *
answer
SDS PAGE
question
proteins are separated based of the isoelectric point exploits the acidic and basic properties of amino acids by separating on the basis of pI placed into a gel with a pH gradient (acidic is anode/ basic is cathode) electric field is then generated across the gel proteins with + charge go to the cathode proteins with - charges go to the anode as the protein that reaches the proportion of the gel where pH is equal to the protein's pI - the protein will stop moving ex. protein has a pI of 9 - basic when at ph of 9 will have no net charge and will stop moving when pH is seven - there will be more protons around the solution - protons will attach to the basic sites on the protein creating a more net positive charge the net positive charge (cations) will be attracted to the cathodes acids with protons - carry a positive charge - thus the anode is positively charged (anions are negative charges)
answer
isoelectric focusing
question
PI ph at which the protein or amino acid is electrically neutral with equal number of positive and negative charges for polypeptides - this is determined by the relative numbers of acidic and basic amino acids
answer
isoelectric point
question
uses physical and chemical properties to separate and identify compounds from a complex mixture is about the affinity of a substance for the mobile and stationary phases - except for size exclusion chromatography *homogenized protein mixture to be fractioned through a porous mixture* - isolated proteins are immediately available for identification and quantification more SIMILAR the compound is to the surroundings (polarity/charge) the more it will STICK and the SLOWER it will go preferred over electrophoresis when large amount of protein are being separated place sample into the stationary phase mobil phase is run through the stationary phase (this will allow the sample to go through the stationary phase/elute) depending on relative affinity of the sample for the stationary and mobile phases the different substances will migrate at different times high affinity for stationary phase = no movement high affinity for mobil phase = move quickly types: Column ion Exchange Size Exclusion Affinity use of different media as the stationary phase allows us to exploit different properties that allow is to separate the compound
answer
chromatography
question
absorbant solid medium used in chromatography
answer
stationary phase
question
the amount of time a compound spends in the stationary phase in chromatography varying retention times of each compound in the solution results in separation of the components within the stationary phase / partitioning
answer
retention time
question
separation of compounds within the stationary phase due to the varying retention times of each compound
answer
partitioning of compounds
question
stationary phase: silica or alumina beads gravity moves the solvent and compounds down the column both size and polarity have a role in determining how fast the compound goes through the absorbent LESS POLAR = faster it will move/ shorter retention time MORE POLAR = slower it will move/ longer retention time different fractions that drip out correspond to different compounds can separate and collect other macromolecules besides proteins (nucleic acids)
answer
Column Chromatography
question
beads in the column are coated with *charged substances* attract and bind to compounds with opposite charges if beads are coated with negative charges - positive charged proteins will have the longer retention time salt gradient is used to elute the charged molecules that have stuck to the column
answer
ion exchange chromatography
question
beads in the column contain tiny pores of varying sizes the small pores allow small molecules to enter the beads - SLOWING THEM DOWN larger compounds cannot fit into these tiny pores so the move around the beads and are eluted out of the column faster the size of the pores may be varied so the molecules of different molecular weights can be fractioned
answer
size exclusion chromatography
question
customizing columns to bind to any protein creating the column so it has a high affinity to that specific protein coating the beads with a receptor that bind the protein or a specific antibody for that protein - thus the protein is retained in the column common stationary phases: nickel (used in separation of genetically engineered proteins with histidine tags - antibodies/ antigens -enzyme substrate analogues (mimic the natural substance for the enzyme of interest) once the protein is retained - it is eluted with a free receptor which will compete with the bead bound receptor and ultimately free the protein from the column elutes can be specific in pH or salinity which will disrupt the bonds associated with the receptor bound to the bead drawback: recovered substances can be bound to the elute - and that can be hard to remove
answer
affinity chromatography
question
determined through X ray crystallography and nuclear magnetic resonance NMR spectroscopy
answer
determining protein structure
question
way of determining protein structure most reliable and common method measures electron density on a high resolution scale - can be used for nucleic acids x ray diffraction pattern is generated - the small dots in the diffraction pattern than can be interpreted to determine the proteins structure
answer
X ray crystallography
question
can be determining by complete protein hydrolysis and subsequent analysis - random nature of the hydrolysis prevents the proper sequencing to determine the sequence/ primary sequence Edman degradation larger proteins digestion of chymotrypsin, trypsin, cyanogen bromide - cleaves specific amino acid residues - creating smaller fragments that can be analyzed by electrophoresis or the Edman degradation salt bridges cannot be determined by these methods
answer
determining amino acid composition
question
determined by monitoring a known reaction with a given concentration of substrate and comparing it to the standard activity is correlated with concentration can be affected by purification methods and conditions of the assay color analysis can determine concentration
answer
determining protein activity
question
cleaves proteins to sequence them (up to 50-70 amino acids) electively and in order removed the N terminal amino acids then can be analyzed via mass spectroscopy
answer
edman degradation
question
concentration is determined through SPECTROSCOPY aromatic side chains - UV spectroscopy proteins can cause color changes in BCA assay, Lowry reagent assay and Bradforf protein assay amount of color change corresponds to concentration
answer
determining protein concentration
question
assay to determine the concentration of protein in solution protein is put into the solution with Coomassie Brilliant Blue Dye dye gives up protons to the ionizable groups in the protein --> turning it blue non-covalent attractions between the deprotonated die and the protein stabilize this blue form of the dye protonated form = brown green color deprotonated form = blue known protein is but into the assay, and an absorbance is measured to create a standard curve unknown sample is put into the same conditions and then an absorbance curve is measured and compared to the standard curve limited by the presence of detergent or excess buffer
answer
Bradford Protein Assay
question
most basic carbohydrate unit contain alcohols and either aldehydes or ketones can undergo: oxidation/ reduction reactions esterification reactions nuclophilic attack creating glycosides reactions every carbon other than the carbonyl carbon will carry a hydroxyl group hydroxyl group = NUCLEOPHILE carbonyl group = ELECTROPHILE
answer
monosaccharides
question
most simple monosaccharide contain three carbon atoms
answer
trioses
question
monosaccharide with four carbon atoms
answer
tetroses
question
monosaccharide with five carbon atoms
answer
pentoses
question
monosaccharide with six carbon atoms
answer
hexoses
question
carbohydrates that contain an aldehyde group as their most oxidized functional group ex. glyceraldehyde aldehyde carbon/ carbonyl carbon will always be C1 this carbon can participate in *glycosidic linkages*
answer
aldoses
question
carbohydrates that contain a ketone group as their most oxidized functional group ex. dihydroxyacetone keto carbon/ carbonyl carbon not C1! this carbon can participate in *glycosidic linkages* (most ketoses it will be C2)
answer
ketoses
question
most basic monosaccharide triose aldose polyhydroxylated aldehyde carbonyl group is most oxidized - numbered as the lowest number
answer
glyceraldehyde
question
the sugars that are acting as substitutes via this linkage
answer
glycosyl residues
question
simplest ketone sugar - ketotriose carbonyl carbon is most oxidized
answer
dihydroxyacetone
question
stereoisomers compounds that have the same chemical formula *they only differ in terms of their spacial arrangement of their component atoms* # of possible stereoisomers = 2^n n= number of chiral carbons in the molecule types: enantiomers distereomers -- epimers
answer
optical isomers
question
stereoisomers that are non identical, nonsuperimposable mirror images of each other D and L any molecule that contains a chiral center and non internal planes of symmetry has one of these a molecule can only have one of these! has to flip at all the chiral centers
answer
enantiomers
question
three dimensional arrangement of the groups attached to the chiral carbons R/S system used for organic chemistry D/L system used in biochemistry they are not interchangable!!! D sugars = hydroxide on their highest carbon to the right L sugars - hydroxide on their highest carbon to the left 8every chiral center in D will have the opposite configuration of the L and vise versa
answer
absolute configuration
question
cannot be determined due to the L/D absolute configuration needs to be determined experientially
answer
direction of rotation
question
Horizontal lines = wedges/ out of the page vertical lines = dashed/ into the page
answer
Fisher Projection
question
type of stereoisomers/ optical isomers same family, same carbons but are not identical and are not mirror images of each other can be different at many different carbons molecule can have many of these
answer
diastereomers
question
diastereomers that differ in configuration from one another at exactly one carbon (alpha and beta are examples of C1 epimers)
answer
epimers
question
nucleophile (OH) can attack the electrophile (C=O) on the same monosaccharide this cam form cyclic hemiacetals/ hemiketals pyransoses/furanose are the only stable ones due to ring strain - usually they exist in the cyclic forms hydroxyl oxygen become a member of ring regardless of hemiacetal or hemiketal - *the carbonyl carbon becomes CHIRAL*
answer
Cyclic sugar formation via intramolecular nuclophilic addition
question
six membered ring from intramolecular nuclophilic addition
answer
pyransose
question
five membered ring from intramolecular nuclophilic addition
answer
furansose
question
carbon that was the carbonyl carbon always becomes chiral
answer
anomeric carbon
question
molecules that differ at the anomeric carbon these are epimers!
answer
anomers
question
OH group in the C1 is trans to the CH2OH *it will be axial and down*
answer
alpha anomer
question
the OH group of the C1 cis to the CH2OH group *it will be equatorial and up*
answer
beta anomer
question
three dimensional conformation of cyclic structures five cycle - planar six cycle - chair axial and equatorial to minimize steric hindrance down on right of Fisher Up on left on fisher
answer
Haworth projection
question
spontaneous change of conformation about C1 from the beta anomer to the alpha anomer due to exposing the hemiacetal rings to water which will cause them to spontaneously cycle between the open and closed form occurs more rapidly when the reaction is catalyzed with an acid or base result is a mixture that has both anomers at equilibrium 36% is alpha and 64% is beta -* hydroxyl group of the anomeric carbon is axial - adding to the steric strain of the molecule when it is in alpha form*
answer
mutarotation
question
this this one of the most important reactions because it yields energy! as the monosaccharides go through maturation they spend a little time in the open aldehyde/ketone form - opportunity for them to be *oxidized into carboxylic acids* aldoses - reducing agents (because they can be oxidized) reducing sugar to aldonic acids, alditols, or a deoxy sugar
answer
Oxidation of Carbohydrates
question
oxidized aldehydes to carboxylic acids
answer
aldonic acids
question
any monosaccharide with a hemiacetal ring
answer
reducing sugar
question
when a aldose in ring form undergoes a oxidation reaction contains a cyclic ester (lactone) with a carbonyl group on the anomeric carbon ex. vitamin C
answer
lactone
question
uses Ag(NH3)2+ as an oxidizing agent aldehydes reduce Ag+ to metallic silver aldehyde and ketones (through tautomerization) gives positive tests for these
answer
tollens reagent
question
aldehyde group of an aldose is oxidized - which is indicated by the red precipitate of Cu2O aldehyde and ketones (through tautomerization) gives positive tests for these
answer
benedict's reagent
question
only reacts with glucose - no other reducing sugars
answer
glucose oxidase
question
will oxidize both the aldehyde group and the primary alcohol on C6 to carboxylic acids
answer
nitric acid
question
rearrangment of bonds in a compound moves a hydrogen and forming a double bond keto to enol shifts ketone group picks up a hydrogen while the double bond is moved to the adjacent carbons resulting in the enol not as common as mutarotation but still does happen
answer
tautomerization
question
group with a double bond and an alcohol group
answer
enol
question
aldehyde group of an aldose is reduced to a alcohol
answer
alditol
question
contains a hydrogen that replaces a hydroxyl group on the sugar D-2-deoxyribose (carbohydrate found in DNA) was reduced to now have a H instead of a OH
answer
deoxy sugar
question
monosaccharides (because they have hydroxyl groups) are able to work with carboxylic acids and carboxylic acid derivatives (ANYHDRIDES) to form *esters* act as protecting groups similar to the phosphorylation of glucose all the OH on the monosaccharide have the addition of the R group
answer
esterification of carbohydrates
question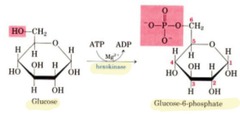
creates a phosphate ester this process is very important reaction in which a phosphate group from ATP is transferred to glucose (and ending with ADP) *hexokinase* *glucoinase* - in liver and pancreatic beta islet cells both enzymes does this reaction

answer
phosphorylation of glucose
question
formation of acetals from hemiactals or formation of ketals from hemiketals glucose with ethanol (or any alcohol) OR group on the alcohol adds to the anomeric carbon and the OH which was on the anomeric carbon forms water creating these bonds is a *dehydration reaction* breaking these bonds is a *hydration reaction*
answer
glycoside formation of carbohydrates
question
formed when hemiacetals react with alcohol the anomeric hydroxyl group is transformed into a alkoky group - making alpha and beta acetals with water as the leaving group
answer
acetals
question
the C-O bond in a acetal that was just formed due to the addition of the alcohol disaccharides and polysaccharides form due to these forming in between monosaccharides creating these bonds is a *dehydration reaction* breaking these bonds is a *hydration reaction* because it can happen at any hydroxyl group - this allows for branching in polysacchrides
answer
glycoside bonds
question
furanose rings connected by glycoside bonds
answer
furanosides
question
pyranose rings connected by glycoside bonds
answer
pyranosides
question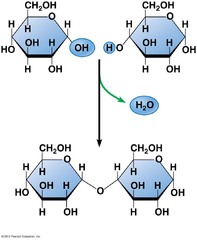
reaction of two hydroxyl groups on two different monosaccharides hydroxyl group of the anomeric carbon reacts with another hydroxyl group of another sugar to form an acetal 1,2 1,4 1,6 glycosidic linkage if between two anomeric carbons both (alpha's/beta's) need to be written with a 1,1 commonly produced in the cell by enzymatic activity

answer
formation of disaccharide
question
important disaccharide *glucose ?-1,2-fructose*
answer
sucrose
question
important disaccharide *galactose-?-1,4-glucose*
answer
lactose
question
important disaccharide *glucose-?-1,4-glucose*
answer
maltose
question
long chains of monosaccharides linked together by glycosidic bonds ex. cellulose, starch, glycogen (all made up of same monosaccharide - D- glucose) differ at the anomeric carbon and the position of the glycosidic bonds
answer
polysaccharides
question
polysaccharide that is composed of only one monosaccharide
answer
homopolysaccharide
question
polysaccharide that is composed of multiple different monosaccharides
answer
heteropolysaccharide
question
happens when an internal monosaccharide in a polymer chain forms at least tow glycosidic bonds
answer
branching in polysaccharides
question
main polysaccharide (homopolysaccharide) made of *?-D-glucose* main structural component of plants *?-1,4 glycosidic bonds* hydrogen bonds hold the polymer chains together for support we cannot digest this because we do not have the enzyme for it - that is why it is fiber - we cannot digest it so it fills us up - draws water into the gut
answer
cellulose
question
main polysaccharide (homopolysaccharide) made of *?-D-glucose* store this as *amylose* *?-1,4 glycosidic linkage AND ?1,6 glycosidic linkage BRANCHES* - every 25 are the branches iodide is a well known reagent because it tests the presence of starch - it fits inside it making the starch-iodide complex
answer
starch
question
main polysaccharide (homopolysaccharide) made of *?-D-glucose* *?-1,4 glycosidic linkage AND ?1,6 glycosidic linkage BRANCHES* different from starch because it has more BRANCHES every 1- molecules instead of 25 highly branched molecule optimizes the energy and makes it more soluble in solution this allows more glucose to be stored in the body branching pattered also allows the enzyme (glycogen phosphorlyase) to cleave at multiple sites simultaneously
answer
glycogen
question
storage unit of starch in plants does not contain the ?1,6 glycosidic linkage BRANCHES only contains the *?-1,4 glycosidic linkage* degraded by ? amylase and ? amylase
answer
amylose
question
type of starch that contains the *?1,6 glycosidic linkage BRANCHES* as well as the *?-1,4 glycosidic linkage* needs debranching enzymes to break this down
answer
amylopectin
question
cleaves amylose randomly along the chain to yield shorter polysaccharides (maltose, glucose)
answer
? amylase
question
cleaves amylose at the nonreducing end of the polymer the end with the acetal to yield maltose
answer
? amylase
question
cleaves glycogen into glucose cleaves from the non reducing end of the glycogen branch and phosphorylates it makes glucose 1 -phosphate
answer
glycogen phosphorlyase
question
insoluble in water soluble in nonpolar organic solvents serve as coenzymes (in electron transport chain and in glycosylation reactions) function as hormones - transmit signals over long distances and as intracellular messengers responding to extracellular signals if have conjugated double bonds - they can absorb light which helps us see others act as pigments types: 1.phospholipids ---glycerophospholipids *glycerol backbone* 2. sphingolipids *sphinosine backbone* ---sphingomyelins (also could be considered phospholipids) ---glycosphingolipids (cerebrosides, globosides) *glycolipids that are 0* ---gangliosides *glycolipids that are -* 3. Terpenes ---steroids 4. postaglandins 5. triacylglycerols
answer
lipids
question
hydrophobic and hydrophilic regions ex. membrane phospholipids and cholesterol are this when placed into a aqueous solution - these molecules spontaneously form structures that allows the hydrophobic regions to group internally the hydrophilic regions interact with water leads to the formation of liposomes, micelles and phospholipid bilayers
answer
amphipathic
question
amphipathic polar head - hydrophilic region fatty acid - hydrophobic region
answer
membranes
question
contain: 1. phosphate and alcohol (polar head group) 2. hydrophobic fatty acid tail liked by a phosphodiester linkage the hydrophobic fatty acids are attached to a backbone to form the hydrophobic fatty acid tail can be classified based on their backbone (glycerol backbone and sphinosine backbone) *tail is always composed of long chain fatty acids* types: *glycero*phospholipids
answer
phospholipids
question
backbone for *phosphoglycerides* and *glycerophospholipids*
answer
glycerol
question
backbone fore *sphingolipids*
answer
sphinosine
question
attach to a backbone to make the hydrophobic fatty acid tail part of the phospholipid hydrocarbons that vary on their degree of saturation and length
answer
fatty acids
question
fatty acid tails that only contain single bonds saturated because it is bonded to four other atoms no pi bonds! in butter - solid at room temperature greater van der Waals forces are are more stable main components in animal fats considered less healthy these decrease the overall membrane fluidity
answer
saturated fatty acids
question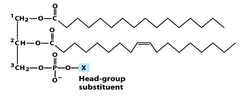
phosphoglycerides are phospholipids that contain a *glycerol backbone* bonded by *ester linages* to *two fatty acids* and by a *phosphodiester linkage* to a highly polar head group named based on their *head group* - because they all have the same backbone head group determined the membrane surface proteins types: phosphatidycholine phosphatidylethanolamine head group can be +, -, 0 membrane surface proteins of these molecules make them very important to cell recognition, signaling and binding

answer
glycerophospholipids
question
type of glycerophospholipid or sphingomyelins with CHOLINE as the head group
answer
phosphatidycholine
question
type of glycerophospholipid or sphingomyelins with ETHANOLAMINE as the head group
answer
phosphatidylethanolamine
question
cell surface antigens are this (ABO antigens) sites of biological recognition at the cell surface and can be bonded to various head groups and fatty acids have a *sphingosine or sphingoid* backbone have long chain nonpolar fatty acid tail with a polar head group can be *phosholipids* (because contain *phosphodiester linkage*) or can be a *glycolipid* (because they contain *glycosidic linkages*) classes: 1. sphingomyelins 2. glycosphingolipids --> cerebrosides 3. ceramide 4. gangliosides 5. glycosphingolipids --> globosides
answer
sphingolipids
question
single hydrogen atoms as its head group sphingolipid
answer
ceramide
question
class of sphingolipids are PHOSPHOLIPIDS - contain a phosphodiester bond have either a *phosphatidylcholine* or *phosphatidylethanolamine* the head groups have *no net charge* major components in the plasma membrane of cells producing *myelin (oligodendtocytes and Schwann cells) in the cell membranes of cells that are used for insulating sheath for axons
answer
sphingomyelins
question
sphingolipids that head groups composed of sugars bonded by *glycosidic linkages* GLYCOLIPIDS found mainly in the outer surface of the plasma membrane two types: 1. Cerebrosides 2. Globosides also known as neutral glycolipids - *no net charge* at a physiological pH
answer
glycosphingolipids
question
type of glycosphingolipids have a single sugar
answer
Cerebrosides
question
type of glycosphingolipids have two or more sugars
answer
Globosides
question
type of sphingolipids (sphingosine or sphingoid backbone) GLYCOLIPIDS glycosidic linages - no phosphate grou; polar head group is oligosaccharides with one or more *N-acetylneuramanic acid (NANA/stalic acid)* at the terminus this contains a *negative charge* play a role in cell interaction, recognition, and signal transduction
answer
gangliosides
question
esters of long chain fatty acids with a long chain alcohol for pliable solids at room temperature secreted as a surface containing to prevent excess evaporation protect against parasites secreted to prevent dehydration water repellant keep skin dry lubricant
answer
waxes
question
odiferous chemicals (chemicals that give off or have a specific smell) - strongly scented class of lipids from *isoprene* metabolic precursors steroids and other lipid signaling molecules made primarily by plants and insects grouped according to the number of isoprene units they present
answer
terpenes
question
monomers than make up terpenes carbons grouped in multiples of five
answer
isoprene
question
made up of terpenes
answer
resin
question
C10H6 two isoprene units abundant in essential oils and turpentine
answer
monoterpenes
question
three isoprene units
answer
Sesquiterpenes
question
four isoprene units ex. vitamin A --> makes retinal (pigment necessary for sight)
answer
diterpenes
question
six isoprene units can be converted to cholesterol and other steroids
answer
triterpenes
question
tetraterpenes - have eight isoprene units ex. beta carotene and lutein
answer
carotenoids
question
isoprene chains between 1000-500 units long ex. rubber
answer
polyterpene
question
derivatives of terpenes *four cycloalkane rings* fused together - three cyclohexane -one cyclopentane the functions of these are determined by the oxidation of these rings along with the functional groups they carry They are NONPOLAR because of the large number of carbons and hydrogens they carry 17 carbons!
answer
steroids
question
steroids that ACT like hormones secreted by endocrine glands into the bloodstream and then travel on protein carriers to distant sites - they can then bind to high affinity receptors and alter gene expression levels regulate gene expression and metabolism - they affect a wide variety of biological systems ex. testosterone, estrogens, cortisol, aldosterone
answer
steroid hormones
question
steroid derivative of a triterprene component of the phospholipid bilayer responsible for mediating membrane fluidity is an amphipathic molecule interacts with the hydrophobic tails and hydrophilic heads of phospholipids - this helps maintain the relative constant fluidity in cell membranes keeps the cell from solidifying at low temperatures also keeps it intact and prevents it from becoming too permeable serves as a precursor to: *other steroid hormones, bile acids, vitamin D*
answer
Cholesterol
question
produced by every cell in the body! 20 carbon molecules unsaturated carboxylic acids (derived from *arachidonic acid* and contain a five carbon ring can act as a paracrine or autocrine molecule regulate the synthesis as cyclic adenosine monophosphate (cAMP) - cAMP than goes on to effect other thinds in the body (smooth muscle function, sleep wake cycle, elevation of body tempature due to pain or fever) COX (cyclooxygenase) enzyme that helps make this - anti-inflammatory drugs inhibits this enzyme
answer
prostaglandins
question
fat soluble vitamin also called *carotene* unsaturated hydrocarbon that is important for vision, growth and development, and immune function aldehyde form - retinal storage form - retinol
answer
vitamin A
question
fat soluble vitamin also called *cholecaliferol* consumed or formed in UV light driven reaction in the skin in liver and kidneys, converted to *calcitrol* (active form of this (uptake of calcium and phosphate in the intestine to promote bone growth) - vitamin D added to milk in order to aid in the absorption of calcium
answer
vitamin D
question
fat soluble vitamin aromatic ring with a long isoprenoid side chain hydrophobic deines tocopherols and tocotrienols
answer
vitamin E
question
fat soluble vitamin group of compounds (K1 and K2) vital in the post-translational modification required to form prothrombin aromatic ring of this undergoes a cycle of oxidation and reduction in the formation of prothrombin is also required to introduce calcium binding sites on several calcium dependent proteins
answer
vitamin K
question
aldehyde form of vitamin A component of the light sensing molecular system in the human eye
answer
retinal
question
storage form of vitamin A can be oxidized to form retinoic acid
answer
retinol
question
oxidized form of retinol hormone that regulated gene expression during epithelial development
answer
retinoic acid
question
active form of vitamin D happens in the liver or kidneys increases calcium and phosphate uptake in the intestines ---> this promotes bone production
answer
calcitrol
question
due to a lack of vitamin D seen in children and characterized by underdevelopment curved long bonds impeded growth
answer
rickets
question
biological antioxidants aromatic ring reacts with free radicals - destroying them prevents oxidative damage - important contributor to the development of cancer and aging
answer
tocopherols
question
K1
answer
phylloquinone
question
K2
answer
menaquinones
question
clotting factor in the blood vitamin K is used to post translationally modify to create this
answer
prothrombin
question
anticoagulant works by blocking the recycling of vitamin K - causing a deficiency or lowering of the active amount of vitamin K should stay away from foods containing high amounts of this vitamin (green leafy vegetables)
answer
warfin
question
called triglycerides used for energy storage composed of three fatty acids bonded by ester linkages to glycerol - rare to see that all three fatty acids are the same nonpolar and hydrophobic (INSOLUBLE IN WATER) both polar components of the glycerol (OH groups) and the free fatty acids (carboxylates) are bonded together - decreasing the polarity look like oil droplets in the cytosol serve as depots of metabolic fuel - recruited to cells that need them adipocytes store large amounts of these - primarily under the skin travel bidirectionally in the blood between the liver and the adipose tissue *hydrolyzed by naturally occurring bases to form salt* for storage of energy 1. carbon atoms of the fatty acids are more reduced than those of sugars because the sugars contain more OH groups - oxidation can yield twice the amount of energy per gram (more energy dense storage mechanism) 2. these are hydrophobic - they do not draw water in and due to require hydration for stability
answer
triacylglycerols
question
unesterified fatty acids with a free carboxylate group circulate in the blood and bond noncovalently to serum *albumin* make up SOAP (through saponification
answer
free fatty acids
question
ester hydrolysis of triglycerides using a strong base (lye) basic cleavage of a fatty acid - leaving the sodium salt of the fatty acid and glycerol fatty acid salt is what we know as salt
answer
saponification
question
base used in saponification common name for sodium or potassium hydroxide
answer
lye
question
lowers the surface tension of the surface of a liquid serves as a detergent or an emulsifer ex. soap two phases originally - add this and then the colloid is formed
answer
surfactant
question
one phase of two liquids occurs because of the formation of micelles
answer
colloid
question
tiny aggregates of soap with all the hydrophobic tails turned inward and the hydrophobic heads are turned outward this shields the hydrophobic lipid tails and allows for overall solvation also important for the absorption of the fat soluble bases and other complicated lipids (lecithins) form in the intestine due to the fatty acids and bile salts secreted by the gallbladder increase the surface area available for lipolyitic enzymes
answer
micelles
question
two forms 1. deoxyribonucleic acid (DNA) 2. ribonucleic acid (RNA) differ at their pentose ring RNA pentose is a ribose DNA pentose is a deoxyribose (when the C2 OH is replaced with a H)
answer
nucleic acids
question
form of nucleic acid *polydeoxyribocuclotide* monodeoxyribonucleo*tides* linked together Bulk of this is found in the chromosomes in the nucleus, also found in mitochondria and chloroplasts deoxyribose (pentose sugar) and phosphate backbone - backbone is composed of alternating sugar and phosphate groups -determines the directionality of DNA - always read from 5' to 3' -nucleotides are formed by 3'-5' phosphodiester bonds (*3' carbon on one sugar to the 5' phosphate group on the incoming sugar (phosphate is one the C5))*
answer
DNA Strand Polarity
question
form of nucleic acid
answer
RNA
question
five carbon sugar/ pentose bonded to a nitrogenous base. nitrogenous base is liked to the C1 of the pentose ring NO PHOSPHATE
answer
nucleosides
question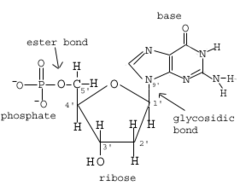
formed when one or more phosphate groups are attached to the C5 carbon of the pentose ring on the nucleoside named according to how many phosphate groups they have ATP, ADP, AMP TTP, TDP, TMP CTP, CDP, CMP GTP, GDP, GMP High energy compounds energy associated with the repulsion between the closely associated negative charged on the phosphate groups *usually bond breaking is endothermic - bond making is exothermic* EXCEPTION: *due to all the negative charges in close proximity removing a terminal phosphate from ATP actually RELEASES ENERGY (exothermic) - powers our cells* THESE ARE THE BUILDING BLOCKS OF DNA

answer
nucleotides
question
OH or phosphate group on the C5 carbon of the deoxyribose
answer
5' end of DNA
question
free OH on C3
answer
3' end of DNA
question
5'----ATG----3' 3'----GTA----5' pApTpG dAdTdG
answer
DNA writing
question
purines and pyrimidines both are *aromatic and heterocycles*
answer
nitrogen containing bases in DNA
question
type of nitrogen containing base in DNA contain two rings in their structure adenine - found in RNA and DNA guanine - found in RNA and DNA
answer
purines
question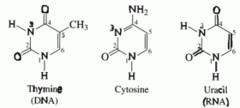
type of nitrogen containing base in DNA contain only one ring cytosine - found in DNA and RNA thymine - found in DNA uracil - found in RNA

answer
pyrimidines
question
unusually stable ring system rules: cyclic planar conjugated (has alternating single and multiple bonds, or lone pairs, creating at least one unhybridized p orbital for each atom in the ring 4n + 2 pi electrons (n is any integer) = Huckel's rule most common is benzene ring - extra stable due to the delocalized pi electrons - delocalized electrons can travel throughout the entire compound using available molecular orbitals -each carbon is sp2 hybridized -each of the six orbitals overlaps equally with its two neighbors - form two pi clouds, one above and one below UNREACTIVE - because they are so stable
answer
aromatic
question
4n+ 2 pi electrons n is any integer rule in making a structure aromatic
answer
Huckel's rule
question
contain at least two different elements in the ring
answer
heterocycles
question
two strands are antiparallel (two strands are going in opposite directions) sugar phosphate backbone is in the outside of the helix with the nitrogen containing bases on the inside specific base pairing rules = complementary base pairing A=T (two hydrogen bonds) G=C (three hydrogen bonds) if denatured it can be reannelaed/brought back together if the recovery process is slowed -important for PCR and the probe DNA
answer
watson- crick model
question
DNA with the known sequence
answer
probe DNA
question
basic proteins DNA is wound around these are basic proteins so they have + charges on it so they attack the - on the phosphate groups of DNA
answer
histones
question
DNA wound around histones
answer
chromatin
question
the DNA wound around the histone (200 bases of DNA) wrapped around H2A, H2B, H3, H4 H1 seals off the DNA as it enters and leaves the nucleosome - adds stability to the structure create a more organized and compacted DNA
answer
nucleosome
question
proteins with nucleic acid prosthetic groups proteins that associate with DNA ex. histones other than histones (basic) they tend to be acidic and stimulate process such as transcription
answer
nucleoproteins
question
chromatin that remains compacted during interphase appears dark transcriptionally *silent* consists of DNA with highly repetitive sequences more methylated - stopping access transcription factors and more to the DNA - silencing the DNA
answer
heterochromatin
question
dispersed chromatin appears light contains genetically active DNA where important genes are located
answer
euchromatin
question
repeating unit of TTAGGG this is at the end of DNA after replication this is shortened can be replaced by *telomerase* shortening of this contributed to aging high GC content created - strong bonds exceptionally strong strand attractions at the end of the chromosomes to prevent unraveling - knotting off the end of the chromosome
answer
telomere
question
enzyme that is conserved in highly dividing cells can replace parts of the telomere
answer
telomerase
question
region found n the center of chromosomes sites of constriction - form noticeable indentations composed of *heterochromatin* repeats that contain *high GC* content during cell division the two sister chromatids can therefore remain connected at the centromere until the microtubules separate the chromatids during anaphase
answer
centromeres
question
replication complex set of specialized proteins that assist the DNA polymerase
answer
replisom
question
DNA unwinds here to begin replication generation of new DNA happens in both directions
answer
origin of replication
question
on either side of the origin of replication because the generation of new DNA goes in both directions
answer
replication forks
question
origin of replication: ONE PER CHROMOSOME - close chromosome - makes two identical circular chromosomes unwinding of DNA double helix: HELICASE -introduces positive supercoils into DNA Stabilization of unwound template strands: SSDNA BINING PROTEINS Synthesis of RNA primer: PRIMASE Synthesis of DNA: DNA POLYMERASE 3 Removal of RNA primers: DNA POLYMERASE 1 (5'-3' EXONUCLEASE) Replacement of RNA with DNA: DNA POLYMERASE 1 Joining of Okazaki fragments: DNA LIGASE removal of positive supercoils ahead of the advancing replication forks: DNA TOPOISOMERASE/ GYRASE - introduce negative supercoils -work ahead of the helicase and cuts one/both strands, relieves torsional pressure and reseals synthesis of telomeres: NOT NEEDED/ DOESNT HAPPEN
answer
Prokaryotic Replication
question
origin of replication: MULTIPLE PER CHROMOSOME -slower than prokaryotes -sister chromatids are made as the replication forks come together and they are held together at the centromere unwinding of DNA double helix: HELICASE -introduces positive supercoils into DNA Stabilization of unwound template strands: SSDNA BINDING PROTEINS Synthesis of RNA primer: PRIMASE -DNA cannot be synthesized is de novo - cannot be paired directly to the parent strand, needs something to hook onto -RNA can be synthesized de novo Synthesis of DNA: DNA POLYMERASE ? AND ? (alpha and delta) Removal of RNA primers: RNASE H (5'-3' EXONUCLEASE) Replacement of RNA with DNA: DNA POLYMERASE ? (delta) Joining of Okazaki fragments: DNA LIGASE removal of positive supercoils ahead of the advancing replication forks: DNA TOPOISOMERASE - introduce negative supercoils -work ahead of the helicase and cuts one/both strands, relieves torsional pressure and reseals synthesis of telomeres: TELOMERASE
answer
Eukaryotic Replication
question
held together at the centromere what is created after the chromosome has gone through replication
answer
sister chromatids
question
enzyme that unwinds DNA this generates two single stranded template strands ahead of the polymerase will cause positive supercoils in the DNA
answer
helicase
question
bind to the unraveled DNA strand (after DNA helicase) and prevent the reassociation of the DNA strand s and the degradation of DNA by *nucleases* purines and pyrimidines are sticky and will want things to hydrogen bond to
answer
ssDNA molecules
question
wrapping of DNA on itself as its helical structure is pushed ever further toward the telomeres during replication
answer
supercoiling
question
produce negative supercoils in DNA to alleviate some of the positive supercoils that helicase put into the DNA they do this by working ahead of the helicase, nicking one or both strands this allows for relaxation of the *torsional pressure* then they reseal the cut strands
answer
DNA topoisomerases
question
the replication process one parental strand is retained in each of the two resulting identical double stranded DNA molecules x
answer
semiconservative
question
responsible for reading the DNA template and synthesizing the new daughter strand *reads the template 3' to 5'* *makes the new strand in a 5' to 3'* one strand in orientated in the correct orientation for DNA pol to read it, the other is not DNA pol 3 - prokaryotes DNA pol ? and ? - eukaryotes when the incoming nucleotide is added, phosphodiester bond is made and a *pyrophosphate*/ PPi is released five in eukaryotes ?, ?, ????, ?, ? cannot complete the synthesis of the 5' end - chromosome gets a little shorter
answer
DNA Polymerase
question
strand that is copied in a direction of the replication fork the parental strand has a polarity of 5' to 3' DNA pol cannot simply read and synthesize this strand can only synthesize in a 5' to 3' direction from a 3' to 5' template okazaki fragments are made as the replication fork continues to move forward, it clears additional space that DNA polymerase must fill in
answer
lagging strand
question
made due to the synthesis of the lagging strand the DNA pol is only capable of synthesizing from a 5' to 3' direction from a 3' to 5' template
answer
okazaki fragments
question
synthesizes RNA primer (10 nucleotides) in the 5' to 3' direction to start replication constantly adding to the lagging strand because the okazaki fragments need to start with a primer
answer
primase
question
removes the primer in prokaryotes also adds new nucleotides to where the primer was
answer
DNA POl 1
question
removes the primer in eukaryotes
answer
RNase H
question
synthesizes the new DNA strand with ? also adds nucleotides to where the primer was works with DNA pol ? are are assisted with PCNA protein to assemble the sliding clamp works in eukaryotes
answer
DNA pol ?
question
helps strengthen the interaction between the DNA polymerases and the template strand polymerase in eukaryotes
answer
sliding clamp
question
works with DNA pol ? to synthesize both leading and lagging strands polymerase in eukaryotes
answer
DNA pol ?
question
works with DNA pol ? and thought to work with DNA repair polymerase in eukaryotes
answer
DNA pol ?
question
replicates mitochondria DNA polymerase in eukaryotes
answer
DNA pol ????
question
works with DNA pol ? on DNA repair works with DNA pol ? and PCNA proteins to assemble the sliding clamp polymerase in eukaryotes
answer
DNA pol ?
question
cells that proliferate excessively because they are able to divide without stimulation from other cells and are no longer subject to the normal controls on cell proliferation can migrate by metastasis
answer
cancer cells
question
migration to distant tissues by the bloodstream or lymphatic system cancer cells can do this
answer
metastasis
question
mutated genes that cause cancer these genes that encode cell cycle related proteins the abnormal alleles encode proteins that are more active than normal proteins - they promote rapid cell cycle advancement *a mutation in ONE copy is sufficient to promote tumor growth - DOMINANT*
answer
oncogenes
question
oncogenes before they are mutated ex. src (sarcoma)
answer
proto genes
question
category of connective tissue cancers
answer
sarcoma
question
ex. p53 pr Rb (retinoblastoma) encode proteins that inhibit the cell cycle or participate in DNA repair processes they normally function to stop tumor progression antioncogens mutations of these genes result in the loss of tumor suppression activity - and thus, promote cancer *inactivation of BOTH alleles is necessary - because if have one good allele than one copy of the normal protein can inhibit the formation of a tumor - RECESSIVE*
answer
tumor suppressor genes
question
tumor suppressor genes
answer
antioncogens
question
DNA pol has this ability in S (synthesis) phase of the cell cycle when incorrect pairing they hydrogen bonding is unstable instability is detected by the DNA pol as the DNA passes through it the base is excised and replaced with the correct one discriminates between parent and daughter strands through methylation parent is more methylated DNA ligase lacks this - so lagging strand has more errors than the leading strand
answer
proofreading
question
in the *G2 phase* of the cell cycle enzymes are encoded by the genes *MSH2 and MLH1* these enzymes detect and remove errors than were placed in during replication and *were missed in the S phase* *these enzymes are homologues to MutS and MutL* - serve a similar function in prokaryotes
answer
mismatch repair
question
NER G1 and G2 eliminates *thymine dimers* recognizes the lesion due to the bulge in the strand *excision endonuclease* makes a nick in the phosphodiester backbone on both side of the thymine dimer and removes it *DNA polymerase* can fill the gap by synthesizing it in the 5' to 3' direction undamaged strand acts as the template nick is sealed by *DNA ligase*
answer
nucleotide excision repair
question
G1 and G2 when the wrong base is in there cysteine deamination - cytosine becomes uracil can be due to a absorption of thermal energy uracil should not be found in DNA *glycosylase enzyme* detects and removed the base - leaving a AP site AP site is recognized by the *AP endonuclease* DNA pol and ligase fill the gap and seal it there mistakes are not large enough to distort the helix - unlike nucleotide excision repair
answer
base excision repair
question
created by UV light formation of a dimer between two adjacent thymine residues this interferes with DNA replication and normal gene expression distorts the shape of the helix
answer
thymine dimers
question
apurinic/apyrimidinic site abasic site - no base! where a base is left open due to Base excision repair recognized by the AP endonuclease
answer
AP site
question
removes the damaged sequence from DNA
answer
AP endonuclease
question
allows a DNA fragment from any source to be multiplied by either gene cloning or PCR provides a means on analyzing and alternating genes and proteins also gives us the reagents necessary for genetic testing (carrier detection, prenatal diagnosis of genetic diseases) useful in gene therapy can give us a specific protein in unlimited quantities (like insulin for diabetics)
answer
Recombinant DNA technology
question
detecting heterozygote status for a particular disease
answer
carrier detection
question
way of creating large amounts of a desired sequence usually first presented in a small quantity or in a heterogenous mixture (where it is with other DNA sequences) goal: to produce large amounts of a homologous DNA for other applications the person needs to ligate (connect) the DNA of interest to a vector/ piece of nucleic acid the recombinant vector (vector + DNA) is placed into a bacteria bacteria is grown into colonies - make sure you get all the bacteria to have the recombinant vector by treating it with antibiotics (bacteria without the vector would be killed off and plasmid contains the DNA of interest) bacteria can then express the gene by making the protein (and person collect that protein) or can lyse and isolate the recombinant vectors the vectors can the be processed be restriction enzymes to release the cloned DNA from the vector
answer
gene cloning
question
nucleic acid (vector) with the DNA of interest ligated to it also needs to make sure that this plasmid has antibiotic resistance
answer
recombinant vector
question
nucleic acid that the DNA of interest will be ligated to usually a bacteria or virus *plasmid* that can be transferred into a bacteria after the DNA is ligated to it needs its own origin of replication
answer
vector
question
restriction endonucleases enzymes that recognize specific double stranded DNA sequences sequences are palindromic (GAATTC) (CTTAAG) can cut through the backbone of the double helix - they have off set cuts - making sticky ends used both on the DNA of interest that you want to insert into the vector and the vector itself
answer
restriction enzymes
question
large collections of known DNA sequences could equal to the genome of an organism DNA fragments are cloned through vectors and utilized for further study can be made of genomic DNA or cDNA
answer
DNA libraries
question
contain large fragments of DNA and include exons and introns uses *restriction endonucleases and DNA ligase to make the library* promoter and enhancer present can not be reliably used for recombinant proteins, transgenic animals or to identify disease causing mutations
answer
genomic DNA libraries
question
complementary DNA these are constructed by reverse transcribing processed mRNA lacks the introns uses *reverse transcriptase and DNA ligase* to make the library promoter and enhancer not present only includes the genes that expressed in the tissue from which the mRNA was isolated genes may by chance be split into multiple vectors - only these libraries can be used reliably sequence specific genes and *identify disease causing mutations, making of recombinant proteins (insulin and vaccines) and produce transgenic animals*
answer
cDNA libraries
question
cDNA library where it only includes genes that are expressed in the tissue from which the mRNA was taken from
answer
expression libraries
question
joining of complementary baise pairing sequences DNA- DNA or DNA- RNA recognition uses two single stranded sequences huge part of PCR and southern blotting
answer
hybridization
question
polymerase Chain reaction automated process that can produce millions of a DNA sequence without amplifying the DNA in the bacteria requires *primers* which are complementary to DNA and flanks the gene of interest (dATP,dCTP,dGTP, dTTP) requires a *DNA polymerase* - need the DNA pol in Thermus aquaticus because the DNA pol in humans doesn't work at high temperatures needs heat to melt the DNA helix apart DNA of interest is denatures, replicated and then cooled slowly to allow the proper annealing of the daughter strands repeated - each time double the amount
answer
PCR
question
used to separate macromolecules by size and charge for DNA - all are negative due to the phosphate groups in the backbone - they will migrate toward the ANODE of an electrochemical cells - preferred gel is the *agrose gel* -longer DNA sequence is the slower it will move often used while doing a southern blot
answer
gel electrophoresis
question
used to detect the precess and quantity of various DNA strands in the same sample DNA cut by restriction enzymes and separated by size through gel electrophoresis still needing to maintain their separation, they are transferred to a membrane then probed wit many copies of ssDNA sequences probe will bind to complementary base pairing - forming a double strand probes are labeled with radioisotopes or other indicator proteins which then can determine the sequence
answer
southern blot
question
uses: template DNA primers DNA pol four different d?TP = deoxyribonucleotides triphosphates four different modified bases: dideoxyribonucleotide = dd?TP once dd?TP incorporated - DNA pol can no longer add to it sequences terminate with the modified base fragments are then separated by gel electrophoresis
answer
DNA sequencing
question
dd?TP contain a hydrogen at C3 rather than a hydroxyl group once the modified bases are incorporated - DNA pol can no longer add to the chain
answer
dideoxyribonucleotide
question
offers potential cures for individuals with inherited diseases made for diseases where the given gene is mutated or inactive - which will give rise to the pathology transferring of the correct gene into the sequence - fixing the disease! example: severe combined immunodeficiency SCID mutation in the gene encoding the ???? chain on interleukin receptors by placing the correct gene into a virus - it can transmit the correct gene to all the human cells for it to reliable: there needs to be efficient gene delivery by the vectors to deliver the cloned gene into the target cells most of the vectors are modified viruses - so they can infect but not complete their life cycle risks: may activate a oncogene if insert into the proto gene
answer
gene therapy
question
altered in their germ line by introducing a cloned gene into a fertilized ova/ embryonic stem cells cloned gene = transgene can be used to study the disease process from early embryonic development through adulthood way of creating one: *fertilized ovum* cloned gene may be micoinjected into the nucleus of a newly fertilized ovum -rarely! the gene will be incorporated into the DNA nucleus of the zygote ovum is then implanted into the surrogate mother hopefully the offspring will contain the transgene in all their cells - including their germ cells and gametes! they can give it to their off spring - effecting multiple generations - coexist with the animals own copies of the gene helpful for studying *dominant effects* transgenic mice may have different numbers of copies of the transgene number of copies of the gene that are inserted cannot be controlled *embryonic stem cells* transgene is inserted into the embryonic stem cell stem cells are the injected into the blastocysts and implanted into the surrogate mother two types of stem cells original stem cell with transgene creates a CHMERA patches of cells - representing the two different stem cells they can be breed to be heterozygous for the transgene or homozygous for the transgene
answer
transgenic mice
question
cloned gene - disease producing allele which is inserted into the fertilized ova or embryonic stem cell to create a transgenic mouse can be used to study the disease process from early embryonic development through adulthood
answer
transgene
question
a gene has been intentionally deleted mice will be models for human disease
answer
knockout mouse
question
genetic code to translate this genetic information into proteins
answer
central dogma of molecular biology
question
unit of DNA that encodes a specific protein or RNA molecule through transcription and translation that unit can be expressed
answer
gene
question
mRNA messenger of genetic information carries information specifying the amino acid sequence of the protein to the ribosome transcribed from the DNA template strand by *RNA polymerase enzymes* happens in the nucleus needs to go through posttranscriptional modifications for it to leave the nucleus its what contains the codons
answer
messenger RNA
question
three nucleotide segments on *mRNA* - mRNA will be the sequences in the box translated into an amino acid 64 of them - 61 for aa's and three for stop codons also referred to as the *reading frame* all written 5' to 3' each is specific for only and only one amino acid there could be many of them for the same amino acid though recognized by the complementary anticodon on tRNA
answer
codons
question
each mRNA molecule translates into only one protein product eukaryotes
answer
monoscistronic
question
starting the process of translation at different locations in the mRNA can result in different proteins in prokaryotes its because the operons contain multiple genes
answer
polycistronic
question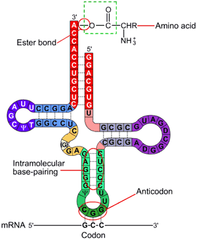
tRNA responsible for converting the language of nucleic acids to the language of amino acids/peptides each contains a a folded strand that includes a three nucleotide anticodon contains a 5' end with a phosphate group contains a 3' end with OH group that contains the activated amino acid attached (ACC nucleotide) charged or activated when the contain an amino acid attached found in the *cytoplasm* attachment of the amino acid to this is a high energy bond amino acids and ATP --> AMP + PP to make the tRNA charged

answer
Transfer RNA
question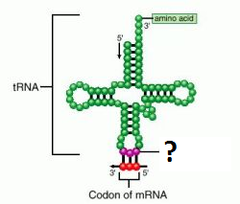
in the folded part on the tRNA recognizes and pairs with the appropriate codon on the mRNA molecule while in the ribosome each amino acids ins represented by at least one codon base pairing is involved - orientation must be anti parallel tRNA - 5' GAU 3' mRNA -5' AUC 3' will be identical to the template strand of DNA

answer
anticodon
question
activates a tRNA by adding a amino acid to it requires to high energy bonds from ATP ATP--> AMP - this bond will be used to supply the energy needed to create a peptide bond during translation
answer
aminoacyl tRNA synthase
question
on the tRNA molecules where the amino acid binds
answer
CCA nucleotide sequence
question
rRNA made in the nucleolus takes place in the ribosomal machinery used during translation in the cytoplasm function as *ribozymes* helps catalyze the formation of the peptide bonds important in splicing out its own intron in the nucleus
answer
Ribosomal RNA
question
enzymes made of RNA molecules instead of peptides many rRNA function as these
answer
ribozymes
question
UGA UAG UAA there are no charged tRNA molecules that can recognize these codons this leads to the release of the protein from the ribosome
answer
stop codons
question
AUG codes for methionine
answer
start codon
question
describing the genetic code because more than one codon can specify a single amino acid amino acids that are encoded for by more than one codon (besides methionine and tryptophan) allows for mutations in the DNA that do not always result in altered protein structure/function
answer
degenerate
question
third position of the codon two bases are usually the same - the third base in the codon is usually variable this is an evolutionary development used to protect against mutations in the coding regions of DNA mutations in this position tend to be *silent or degenerate* - there is no effect on the expression of the amino acid and therefore no adverse effects on the polypeptide sequence
answer
wobble position
question
mutation that occurs and it affects one of the nucleotides in a codon
answer
point mutation
question
point mutations that can effect the first or second nucleotide in the codon - changing the primary amino acid sequence of the protein can be a missense or nonsense mutation
answer
expressed mutations
question
expressed point mutation mutation where one amino acid substitutes for the other
answer
missense mutation
question
expressed point mutation mutation where the codon now encodes for a premature stop codon also known as a *truncation mutation*
answer
nonsense mutation
question
occurs when a number of nucleotides are added or deleted from the mRNA sequence the insertion/deletions will cause a shift in the reading frame this will cause changes in the amino acid sequence or cause a premature truncated protein usually more serious than point mutations
answer
frameshift mutations
question
caused by a frameshift deletion in the 508 codon of the genetic code that codes for the CFRT chloride channel loss of the phenylalanine channel will cause a defective chloride channel the protein/channel will never reach the membrane - water and salt can not go in and out of the cells cells that line the lungs, pancreas and other organs secrete an abnormally thick sticky mucus that traps bacteria and increases the likelihood of infection
answer
cystic fibrosis
question
mRNA fro the DNA template produces a copy of only one of the two strands of DNA helicase and topisomerase are involved in unwinding the DNA and preventing the formation of supercoils results in a *single strand of mRNA* synthesized from the template strand RNA pol 2 binds to the TATA box at -25 first nucleotide is transcribed at +1 stops when it reaches the termination sequence DNA double helix reforms hnRNA is formed mRNA derived from hnRNA via posttranscriptional modifications
answer
transcription
question
DNA region which binds to the DNA dependent RNA polymerase ex. TATA box
answer
promoter
question
main DNA dependent RNA polymerase that is the main player in transcribing mRNA binds to the TATA box/ promoter region located in the nucleus *makes hnRNA and smaller nuclear RNA (snRNA)* does not require a primer to start generating a transcript (unlike DNA pol three in DNA replication)
answer
RNA polymerase 2
question
promoter region in which RNA polymerase 2 binds to named for it high concentration of thymine and adenine bases -25
answer
TATA box
question
help the RNA polymerase locate and bind to the promoter region of DNA helps to establish where the transcription will start *have to be made and translocated back to the nucleus - they are called TRANS regulators because they have to travel through the cell to their point of action* transcriptional activating proteins search the DNA for the specific DNA binding motifs two domains: DNA binding domain activation domain
answer
transcription factors
question
located in the nucleus and synthesizes rRNA transcribes the 28S, 18S and 5.8S rRNA in the eukaryotic ribosome as a single unit in the nucleolus *results in a 45S ribosomal precursor*
answer
RNA polymerase 1
question
located in the nucleus synthesizes tRNA and some rRNA
answer
RNA polymerase 3
question
three in eukaryotes RNA pol 1 RNA pol 2 RNA pol 3 travels along the template strand in the 3' to 5' direction constructs the mRNA in the 5' to 3' direction does NOT DO PROOFREADING
answer
DNA dependent RNA polymerase
question
sense strand not used as a template during transcription since it is complementary to the template strand it is *identical* to the mRNA but thymine nucleotides in this strand have been replaced with uracil
answer
coding strand
question
the first base that is transcribed from DNA to RNA
answer
+1
question
upstream toward the 5' end given negative numbers
answer
left of +1
question
downstream toward the 3' end given positive numbers
answer
right of +1
question
primary transcript heterogenous nuclear RNA forms into mRNA after posttranscriptional modifications
answer
hnRNA
question
needs to happen before the hnRNA can leave the nucleus must undergo three things before it can interact with the ribosome: 1. intron/exon splicing 2. 5' cap 3. 3' poly end tail
answer
post-transcriptional processing
question
post-transcriptional modification removes the noncoding sequences/ introns and ligates the exons/coding sequences together accomplished by the spliceosome introns are taken out as a lariat then degraded
answer
splicing of exon and introns
question
snRNA molecules couple with snRNP's snRNP/snRNA complex recognizes the 5' and the 3' end of the splice sites on the introns
answer
spliceosome
question
snurps bind to snRNA's to form the spliceosome small nuclear ribonucleoproteins
answer
snRNP
question
lasso shaped structure introns are taken out this way then degraded
answer
lariat
question
in the hnRNA and not in the mRNA non coding regions play an important role in the regulation of cellular gene expression levels maintain the size of the genome existence of these has also allowed for rapid protein evolution - many organisms share common peptide sequences suggests that the genes encoding for these particular peptides may employ a modular function - they contain standard sequences that can be swapped in and out depending on need
answer
introns
question
post-transcriptional modification at the end of the 5' part of hnRNA the *7-methylguanylate triphosphate cap* is added added during the process of transcription and recognized by ribosomes as the binding site protects mRNA from degradation in the cytoplasm
answer
5' cap
question
polyadenosyl tail added to the 3' end protects against rapid degradation fuse for the time bomb as leaves the nucleus will get degraded from the 3' end - long the tail is the longer it has to be translated also helps in the export of the mature mRNA from the nucleus
answer
3' poly A tail
question
untranslated regions of mRNA still exist in the 5' to 3' edges of the transcript because the ribosome initiates translation at the start codon (AUG) and will end at one of the three stop codons
answer
UTRs
question
when the primary transcript of the hnRNA may be spliced together in different ways to produce multiple variants of proteins encoded by the same original gene helps to make many more different proteins from a limited number of genes
answer
alternative splicing
question
mRNA can travel from the nucleus to the cytoplasm through this
answer
nuclear pores
question
starts when the mRNA finds the ribosome converts the mRNA transcript into a functional protein requires: mRNA tRNA ribosomes amino acids energy (form of GTP) occurs in the cytoplasm of both prokaryotes and eukaryotes prokaryotes: ribosomes start to translate before the mRNA is complete 50S + 30S = 70S subunit eukaryotes transcription and translation occurs at separate times and locations 60S + 40S = 80S subunit happens in: initiation elongation termination
answer
translation
question
composed of proteins and rRNA large and small subunits - they bind together during protein synthesis three binding site: A, P, E different for between the prokaryotes and eukaryotes - site for antibiotics
answer
ribosome
question
50S and 30S subunits - 70S complete ribosome
answer
prokaryotic ribosome
question
contains for strands of rRNA (28S, 18S, 5.8S, 5S rRNA) genes encoding these strands/rRNA's are found in the nucleolus 45S encoded on the nucleolus (transcribed by RNA pol 3) makes the 45S pre-rRNA becomes the: 1. 18S rRNA of the 40S (small ribosomal subunit) 2. 28S and 5.8S rRNA of the 60S subunit/ large RNA pol 3 also transcribed the 5S rRNA and found in the 60S large subunit two subunits 40S small 60S large join during protein synthesis to form the whole 80S ribosome
answer
eukaryotic ribosome
question
A site in the ribosome holds the incoming aminoacyl- tRNA complex this is the next amino acid that is added to the growing chain determined by the mRNA codon with the A sire
answer
Aminoacyl site
question
P site in the ribosome holds the tRNA that carries the growing polypeptde it is also where the first amino acid (methionine) binds as it is starting the polypeptide chain
answer
peptidyl site
question
E site in the ribosome where the inactivated/ uncharged tRNA passes through before exiting the ribosome since the tRNA is uncharged it quickly unbinds and goes to get recharged
answer
Exit site
question
becomes the: 1. 18S rRNA of the 40S (small ribosomal subunit) 2. 28S and 5.8S rRNA of the 60S subunit/ large
answer
45S pre-rRNA
question
indicate the size of the strand of the rRNA they are determined experimentally by studying the behavior of particles in the ultracentrifuge numbers of each subunit and each rRNA are not additive because they are based on SIZE and SHAPE - not size alone
answer
S value
question
small ribosomal subunit binds to the mRNA prokaryotes 30S binds to the *shine Dalgarno sequence* in the 5' untranslated region of the mRNA first amino acid - Nformylmethionine eukaryotes the 40S binds to the 5' cap structure on the mRNA first amino acid - methionine initiator tRNA binds to the AUG start codon through base pairing of the anticodon in the P SITE large subunit binds to the small subunit - forming a complete initiation complex large subunit binds with help from the initiation factors (IF)
answer
translational initiation
question
three step cycle repeated for each amino acid added to the protein after methionine ribosome moved in the 5' to 3' direction along the mRNA synthesizes the protein from the amino (N) terminus to the carboxyl (C) terminus A site holds the new tRNA P site where the full peptide is E site is where the tRNA is uncharged and quickly leaves elongation factors assist by locating and recruiting aminoacyl-tRNA along with GTP - while helping to remove GDP once the energy has been used some proteins have signal sequences which are for a particular destination hormones/digestive enzyme go to ER - so the protein can be translated directly into the lumen of the rough ER
answer
translational elongation
question
when the stop codon moved into the A site, the protein release factor (RF) binds to the termination codon this causes a water molecule to be added to the polypeptide chain water molecule allows peptidyl transferase and termination factors to *hydrolyze* the completed peptidyl chain from the final tRNA the polypeptide chain will then be released from the tRNA in the P site the two ribosome subunits will dissociate
answer
translational termination
question
helps the large subunit bind to the small subunit they are not permanently associated with the ribosome
answer
initiation factors
question
formed as the polypeptide is passed from the tRNA to the P site to the tRNA in the A site requires *peptidyl transferase* GTP is used for energy during the formation of this bond
answer
peptide bond
question
enzyme that is apart of the large subunit enzyme that is needed to make the peptide bond
answer
peptidyl transferase
question
the protein needs modifications for proper folding chaperones - help in the folding process also modified by cleavage events ex. insulin - cleaved from a larger inactive form to its smaller active form the signal sequence must be cleaved if the protein is to enter the organelle and accomplish its function if they have a quaternary structure the subunits have to come together to form the functional protein ex. hemoglobin phosphorylation carboxylation glycosylation prenylation
answer
post-translational processing
question
specialized class of proteins they assist in the proper folding process
answer
chaperones
question
addition of phosphate by protein kinases to activate or deactivate proteins post-transcriptional modification
answer
phosphorylation
question
addition of a carboxylic acid group usually to serve as calcium binding sites post-transcriptional modification
answer
carboxylation
question
addition of oligosaccharides as proteins pass through the ER and golgi apparatus to determine cellular destination post-transcriptional modification
answer
glycosylation
question
addition of lipid groups to certain membrane bound enzymes
answer
prenylation
question
due to the operon allows to regulate gene expression based on food sources that are available in the enviroment - by sharing a single common promoter region of the DNA sequence (operon) the genes are transcribed as a group
answer
control of gene expression in prokaryotes
question
cluster of genes transcribed as a single mRNA contain structural genes, operator site, promoter site and a regulatory gene two types: inducible repressible
answer
operon
question
described the structure and function of operons
answer
Jacob Monod Model
question
component of the operator in the Jacob Monod Model codes for the protein of interest
answer
structural gene
question
component of the operator in the Jacob Monod Model upstream of the structural gene nontranscribable region of DNA that is capable of binding a repressor protein
answer
operator site
question
component of the operator in the Jacob Monod Model upstream of the operator site provides a place for RNA polymerase to bind
answer
promoter site
question
component of the operator in the Jacob Monod Model most upstream codes for a repressor protein
answer
regulatory gene
question
repressor is bonded tighly to the operator system and acts as a *roadblock* RNA pol is unable to get from the promoter site to the structural gene because the repressor is in the way to remove the block - the inducer must bind to the repressor protein so that the RNA pol can move down the gene as the inducer increases - will pull more copies of the repressor off the operator region - freeing up the genes for transcription the gene products are only made when they are needed ex. lac operon
answer
inducible operon system
question
allow the constant production of a protein product repressor is made by the regulator gene is inactive until it binds to the corepressor the corepressor and repressor binds to the operator site to prevent further transcription tend to be a negative feedback - final protein structure is the corepressor - as levels increase, it can bind to the repressor and the complex will bind to operator and stop transcription ex. trp operon
answer
repressible operon system
question
binding of a protein reduces the transcriptional activity - repressor in the inducible operon system
answer
negative control
question
example of a negative inducible system bacteria can digest lactose - but more energy is needed than digesting glucose only want to digest lactose when *lactose is high and glucose is low* induced by the presence of lactose falling levels of glucose cause an increase in cAMP, which binds to CAP - creating a conformational change, and now can bind to the promoter region - transcription!
answer
lac operon
question
catabolite activator protein transcriptional activator signals that other carbon sources should be used when glucose levels are low cAMP binds to this when glucose levels are low which causes a conformational change with the conformation change it can bind to the promoter of the lac operon - transcription of the lac operon genes
answer
CAP
question
systems in which the binding of a molecule increases transcription of a gene ex. CAP with lac operon
answer
positive control mechanism
question
binds with the repressor together they bind to the operator site and stop further transcription serve as the negative feedback - the final structural protein product is usually this
answer
corepressor
question
negative repressible system when tryptophan is high in the local enviroment it acts as the corepressor binding of the repressor with tryptophan will cause the repressor to bind to the operator site they cell will stop making its own tryptophan will stop wasting energy making their own tryptophan because it is easily available in the enviroment
answer
trp operon
question
transcriptional factors (TRANS regulators) gene amplification in response to specific signals (hormones, growth factors, and other intracellular signals) transcription factors with histones promoters - enhancers and response elements are CIS regulators - because they are in the same vicinity as the gene they control gene duplication
answer
control of gene expression in eukaryotes
question
binds to specific nucleotide sequences in the promoter region or to a DNA response element to help recruit the transcriptional machinery
answer
DNA binding domain on transcriptional factors
question
allows for the binding of several transcriptional factors and other regulatory proteins (RNA pol, histone acetylases - which function in the remodeling of the chromatin structure)
answer
activation domain on transcriptional factors
question
a sequence of DNA that binds only to a specific transcriptional factor DNA binding domain on the transcriptional factor binds to this
answer
DNA response element
question
several response elements allows for the control of ones gene expression by multiple signals transcription factors that bind to their specific response element in the enhancer transcription factor---> response element cAMP--> CREB (cyclic AMP response element binding protein) cortisol --> cocorticoid receptor estrogen --> estrogen receptor there is a large distance between this and the promoter regions - the DNA must often bend into a hairpin loop to bring the elements together spatially can be up to 10000 base away from the regulation gene can be located within an intron by utilizing these - genes have an increased likelihood to be amplified because of the variety of signals that can increase transcription levels
answer
transcription enhancers
question
these are elements that are outside the normal promoter and can be recognized by specific transcription factors that enhance the transcription levels they can be grouped together to form an enhancer
answer
response elements
question
cells can increase the amount of gene product by duplicating the gene genes can be duplicated ins series on the same chromosome - many copies in a row of the same genetic information genes can also be duplicated in parallel - by opening the gene with helicase and permitting DNA replication only of that gene - continue replication until the gene has hundreds of copies of the gene exist in parallel on the same chromosome
answer
gene duplication
question
proteins that are involved in chromatin remodeling can be recruited due to transcription factors they acetylate (CH3C=O) lysine residues found on the N terminal of the histone tail region acetylation of the histone proteins decreases the positive charge on the lysine residues and weakens the interaction between DNA and the histone proteins - the chromatin starts to open and allows for easier access of the transcription machinery to the DNA it increases the space between the histones, allowing for better access to DNA for other transcription factors
answer
histone acetylases
question
proteins that function to remove the acetyl groups from histones make the chromatin in a more closed conformation overal decreases the gene expression levels of the cell
answer
histone deacetylases
question
involved in chromatin remodeling also involved in the regulation of gene expression levels in the cell does this through DNA methylase during development is involved in silencing of genes that no longer need to be activated heterchromatin - more heavily methylated - limits the access of the transcriptional machinery to the DNA
answer
DNA methylation
question
adds a methyl group to cytosine and adenine nucleotides
answer
DNA methylase
question
described as a semipermeable phospholipid bilayer describes function and structure! - semipermeable = chooses which particles can enter and which ones cannot (determined by channels and the membrane itself) phospholipid = two layers on phospholipids permits fat soluble compounds to cross easily larger, water soluble compounds need to go in another way *compared to a capacitor due to having opposite charges on either side* -due to Na K ATPase and Leak channels mostly made of phospholipids with very few free fatty acids - also have steroid molecules and cholesterol which help the fluidity of the membrane - also have waxes with help with membrane stability referred to as the *fluid mosaic model* - in a state of constant flux functions as a stable semisolid barrier between cytoplasm and the enviroment contains: lipid rafts carbohydrates making a glycoprotein coat maybe a cell wall associated with it function: protect the interior of the cell from the external enviroment they selectively regulate traffic into and out of the cell involved in intracellular and intercellular communication and transport contain proteins embedded into the lipid bilayer that act as cellular receptors during signal transduction - the proteins that are embedded play an important role in regulating and maintaining the overall cellular activity phospholipids will move rapidly through out this by simple diffusion (tag them and all of a sudden the tags soon become mixed)
answer
cell/ plasma membrane
question
collections of similar lipids with or without proteins they are attachment points for other molecules serve roles in signaling move with the membrane but more slowly than the phospholipids
answer
lipid rafts
question
created by carbohydrates associated with the plasma membrane the carbohydrates are attached to protein molecules on the extracellular surface because the carbohydrates are hydrophilic they are interact with glycoproteins and water creating the coat
answer
glycoprotein coat
question
primary component in the cell/ plasma membrane they move rapidly in the plane of the membrane through simple diffusion can move in between the layers but is energetically unfavorable because the polar head group of this has to go through the nonpolar tail region of the interior part of the membrane - need flippases to go in between serve as secondary messengers in signal transduction the phosphate group provides as an attachment point for the water soluble groups these are triglycerides but one fatty acids is substituted with a phosphate group the polar head group joins the nonpolar tail (forms a glycerophospholipid which is another name) they can spontaneously assemble into micelles or liposomes due to hydrophobic interactions there are used for membrane synthesis they can produce a hydrophobic surface layer on lipoproteins (such as very low density lipoprotein VLDL)
answer
phospholipids in plasma membranes
question
enzymes that help the lipids move from membrane to membrane and go through the interior part of the membrane
answer
flippases
question
carboxylic acids that contain a hydrocarbon chain and a terminal carboxyl group can be saturated or unsaturated two esstential ones: alpha linolenic acid and linoleic acid
answer
fatty acids in plasma membranes
question
also known as triglycerides storage lipids involved in human metabolic processes they have three fatty acid chains esterfied to a glycerol molecule the fatty acids can be saturated or unsaturated
answer
triglycerols in plasma membranes
question
fatty acids with double bonds healthier fats liquid at room temperature double bonds introduce kinks which makes it difficult for them to stack and solidify they make the membrane more fluid most of these come from the diet they are transported as triglycerides from the intestine inside chylomicrons omega numbering system is used for these usually contains double bonds that are in the cis conformation phospholipids that are in the membrane and have more unsaturated fatty acids make up a more fluid membrane
answer
unsaturated fatty acids
question
essential unsaturated fatty acid maintain cell membrane fluidity 18:3 all cis 9,12,15 - structure of the primary precursor for the omega 3 family
answer
alpha linolenic acid
question
essential unsaturated fatty acid maintain cell membrane fluidity 18:2 cis, cis -9,12 precursor for the omega 6 family (contains arachidonic acid)
answer
linoleic acid
question
very low density lipoprotein lipid transporter phospholipids produce a hydrophobic surface layer on these
answer
VLDL
question
do not contain glycerol still simular in structure to glycerophospholipids/ phospholipids contain a hydrophilic region and two fatty acid derive hydrophobic tails the different kinds of these vary in they hydrophilic regions different classes: ceramide sphingomyelins cerebrosides gangliosides
answer
sphingolipids in plasma membranes
question
necessary for the synthesis of all steroids also helps membrane fluidity contains both a hydrophobic region and hydrophilic region the membrane stability is derived from interactions with both the hydrophilic regions and the hydrophobic regions stabilizes adjacent phospholipids *occupies space between the phospholipids - this prevents the crystal structures in the membrane this increases the fluidity at lower temperatures* at high temperatures limits the movement of the phospholipids within the bilayer - which decreases the fluidity and helps hold the membrane intact 20% of the plasma membrane
answer
cholesterol in plasma membranes
question
lipids that are extremely hydrophobic rarely found in the plasma membrane of animals found in the cell membranes of plants composed of long chain fatty acids and a long chain alcohol - this makes the melting point high in these substances can give stability and rigidity within the nonpolar tail of the region most of them serves as a extracellular function in protection or waterproofing
answer
waxes in plasma membranes
question
pass completely through the plasma membrane
answer
transmembrane proteins
question
are associated with only the interior/cytoplasmic or exterior/extracellular surface of the cell
answer
embedded proteins
question
transmembrane proteins and embedded proteins they both have an association with the interior of the plasma membrane this is due to one of the proteins domains being very hydrophobic
answer
intergral proteins
question
bound by electrostatic interactions with the lipid bilayer, especially lipid rafts or to other transmembrane or embedded proteins (like G proteins found in G protein coupled receptors) ex. transporters, channels, receptors
answer
membrane associated peripheral proteins
question
creates the glycoprotein coat can act as signaling and recognition molecules ex. ABO antigens - sphingolipids that differ in the carbohydrate sequence immune system takes advantage and can recognize these differences of carbohydrates
answer
carbohydrates and the plasma membrane
question
transmembrane proteins types: ligand gated ion channels - respond to a specific ligand G protein coupled receptors - participate in biosignaling and involved in different signaling cascades generally protein (some carbohydrates and lipids - mainly in viruses)
answer
membrane receptors
question
they form a cohesive layer via intercellular junctions they provide direct pathways for communication between neighboring cells or between cells and the extracellular matrix generally composed of cell adhesion molecules (CAM)
answer
cell to cell junctions
question
cell adhesion molecules proteins that allow the cell to recognize each other they contribute to proper cell differentiation and development make up cell to cell junctions
answer
CAM
question
allow for direct cell to cell communication found in small bunches together also called *connexon* forms by the alignment and interaction of pores composed of six *connexin* molecules they permit water movement and some solutes between cells proteins usually cannot be transferred between these
answer
Gap junctions
question
prevent solutes and water from leaking into the space between cells by the paracellular route found in epithelial cells act as a physical link between cells as they form a single layer of tissue enough to create a *transepitheial voltage differences* which is from the difference of concentrations on either side of the epithelium to work they must form a continuous band around the cell can limit permeability they are found in the *lining of the renal tubules* - restrict passage of the solutes and water without cellular control
answer
tight junctions
question
bind adjacent cells together by anchoring their cytoskeletons formed by interactions between transmembrane proteins associated wit intermediate filaments inside the cells usually found in the interface between two layers of epithelial tissue
answer
desmosomes
question
main function is to attach epithelial cells to underlying structures, especially the basement membrane
answer
hemidesmosomes
question
nonspontanous process require energy +????G net movement of solute against the concentration - decrease in entropy ex. primary active transport secondary active transport may or may not be effected by temperature
answer
active transport
question
spontaneous process no not require energy - ????G do not require intracellular stores utilize the CONCENTRATION GRADIENT -supplies the energy needed to make the particles move ex. simple diffusion facilitated diffusion osmosis increase as in rate as the temperature increase *they primary thermodynamic motivator in passive transport is INCREASE in ENTROPY/ ????S* go down the concentration gradient will cause an increase in entropy
answer
passive transport
question
type of passive transport substrates move down their concentartion gradient directly across the membrane only particles that are freely permeable to the membrane can do this there is *potential energy* in the chemical gradient and some of that energy leaves as the gradient is utilized to move the particle (ball down a hill)
answer
simple diffusion
question
type of passive transport simple diffusion of water water will move from a region of lower solute concentration to one of higher solute concentration *will go to the area with more salt/less water* water will go from a more dilute solution/higher water concentration down its concentration gradient to a more concentrated solution/lower water concentration usually the solute is impermeable to the membrane so the solutes can't flow through, but the water will still go through
answer
osmosis
question
type of passive transport simple diffusion of molecules that are impermeable to the membrane large, polar or charged molecules the energy barrier is too large for their molecules to cross freely requires integral membrane proteins to serve as channels or transporters happens through carries and channels
answer
facilitated diffusion
question
concentration of the solutes inside the cell is higher than the surrounding solution water will flow in! - cell will swell maybe making the cell lyse/burst
answer
hypotonic
question
the solution outside the cell has a greater solute concentration than the inside of the cell water will flow out of the cell cell will shrivel up
answer
hypertonic
question
the solute concentration is equal on both side of the cell membrane there is still movement of particles prevents the NET of movement of particles
answer
isotonic
question
colligative property the water level will only rise to a point as which exerts a sufficient pressure to counterbalance the tendency of water to flow across the membrane hydrostatic pressure exerted by the water in the solute containing compartment will eventually oppose the influx of water water will flow into the solute containing compartment M= molarity R= ideal gas constant T = absolute temperature i = van't Hoff factor *directly proportional to the molarity of the solution* is maintained against the cell membrane - rather than the force of gravity - if the osmotic pressure created by the solutes is greater than the pressure that the membrane withstands - the cell will lyse thought as a sucking pressure - drawing pressure into the cell
answer
osmotic pressure
question
physical property of solutions that is dependent on the *concentration* of dissolved particles but NOT the *identity* of the particles ex. osmotic pressure vapor pressure depression/Raoult's law boiling point elevation freezing point depression
answer
colligative property
question
i number of particles obtained from the molecule when in solution glucose = 1 (one intact molecule) NaCl = Na+ Cl- = 2
answer
van't Hoff pressure
question
are only open to one side of the cell membrane at any given point walks into the transport protein, stays in the protein as it undergoes a conformational change and then finally dissociates from the substrate binding site these go through a conformational change for a brief moment of time could be in a *occluded state* - where they are not open to either side of the membrane
answer
carriers
question
viable transporters for facilitated diffusion may be in the open or closed conformation open conformation - channels are exposed to both sides of the cell membrane and act like a tunnel
answer
channels
question
uses ATP or another molecule to directly power the transport of molecules across the membrane usually uses a *transmembrane ATPase* maintains the membrane potential of neurons in the nervous system
answer
primary active transport
question
*coupled transport* uses energy to transport particles across a membrane - *but there is no direct coupling with ATP hydrolysis* it harnesses the energy released by one particle going down the electrochemical gradient to drive another particle up its gradient symport - both are going in the same direction antiport - particles are going in different directions kidneys use this - usually driven by sodium to reabsorb and secrete various solutes into and out of the filtrate
answer
secondary active transport
question
form of secondary active transport both particles are going in the same direction
answer
symport
question
form of secondary active transport particles are going in different directions
answer
antiport
question
when the cell membrane invaginates and engulfs material to bring it into the cell material is encased in a vesicle pinocytosis and phagocytosis substrate binding to specific receptors embedded within the plasma membrane will initiate this process
answer
endocytosis
question
type of endocytosis for fluids and dissolved particles
answer
pinocytosis
question
type of endocytosis for solids such as bacteri
answer
phagocytosis
question
when secretory vesicles fuse with the membrane and release the material from inside the cell to the extracellular enviroment important in the nervous system and intercellular signaling (neurotransmitters from synaptic vesicles)
answer
exocytosis
question
difference in electrochemical potential across the cell membranes maintaing this requires energy because the ions may passively diffuse through the cell membrane through simple diffusion over time through *leak channels* - thus *Na-K pump /ATPase* is needed to regulate the concentration of the intracellular and extracellular sodium and potassium ions -chloride ions also participate in establishing this can be determined by the Nernst equation the impermeability of the cell membrane to the ions and the selectivity of ion channels can lead to electrochemical gradient and thus this resting -40 to -80mV through depolarization can go as high as +35mV
answer
membrane potential
question
usually between -40 and -80 mV
answer
resting potential
question
can be used to determine the membrane potential uses the intra and extracellular concentrations of various ions temp is usually 310K faraday's constant = 96,485 C/mol e-
answer
Nernst Equation
question
comes from the Nernst Equation takes into account the relative concentration of each major ion to the membrane potential chloride is inverted relative to the other ions because it has a negative charge
answer
Goldman Hodgkin Katz voltage equation
question
Na+K+ ATPase function: to maintain a low concentration of sodium ions and high concentration of potassium ions inside the cell does this by pumps three sodium ions out for every two potassium ions that are pumped in net movement of positive charges out of the cell - inside = overall NEGATIVE - outside = overall POSITIVE contain leak channels that allow Na and K to go down the concentration gradient they are more permeable to K than to Na (there are more K leak channels than Na channels)
answer
sodium potassium pump
question
two membranes = inner and outer
answer
mitochondria membrane
question
highly permeable due to many large pores the pores allow for the passage of ions and small proteins this membrane completely surrounds the mitochondrial membrane
answer
outer mitochondria membrane
question
has a much more restricted permeability compared to the outer mitochondria membrane contains cristae/ foldings they increase the available surface area for the integral proteins associated with the membrane the proteins are involved in the electron transport chain and ATP synthase encloses the mitochondria matrix *contains a lot of cardiolipin and does NOT contain cholesterol*
answer
inner mitochondrial membrane
question
enclosed by the inner mitochondrial membrane citric acid cycle produces high energy electron carries in here those electron carriers are used in the electron transport chain
answer
mitochondria matrix
question
glucose can go into most cells it is driven by CONCENTRATION independent of sodium (unlike the absorption from the digestive tract) normal concentration in the periphery is from 4-6mM four transporters GLUT 1-4 GLUT 2 and 4 are the most significant (only located on specific cells and are highly regulated)
answer
glucose transport
question
low affinity glucose transporter found on *hepatocytes and pancreatic cells* -liver cells and in the Betal Islet Cells captures the excess glucose primarily for storage when glucose concentration drops below the Km for this transporter (has a high KM - low affinity for glucose - takes a lot of glucose to bind to this transporter) - it will go past the liver and straight into the periphery the liver will only pick up EXCESS glucose and store it - preferably right after a meal when glucose concentration exceeds the KM this transporter and glucokinase serves as the glucose sensor for insulin RELEASE in the Beta Islet cells of the pancreas
answer
GLUT 2
question
found in *adipose tissue* and *muscle* responds to glucose concentration in the peripheral blood responds to insulin rate of glucose uptake by this transporter is increased by insulin insulin makes these transporters go to the surface of the cells and take up glucose via exocytosis km is close to the normal blood sugar levels - 5mM transporter is saturated when blood glucose levels are just a little bit higher than normal - so even when blood sugar levels are high they will take in glucose at the same amount only way to increase the uptake of glucose is make so there are more glucose transporters on the cell - more insulin more transporters but rate of each transporter is still the same when it is saturated! come into the cell = endoytosis come to the surface of cell = exocytosis muscle stores the extra glucose as glycogen adipose tissue requires glucose to form DHAP, which is then converted to glycerol phosphate to store the incoming fatty acids as triglycerides
answer
GLUT 4
question
irreversible enzyme only found in the liver and pancreatic cells enzyme that is induced by insulin when in the liver enzyme that has a high KM - acts on glucose proportionally to its concentration does the same thing as hexokinase in glycolysis serve as the sensor for insulin along with GLUT 2 in the Beta Islet cells of the pancreas
answer
glucokinase
question
insulin is absent - cannot stimulate GLUT 4 to go to the cell increased glucose concentration in the blood
answer
type 1 diabetes
question
GLUT 4 becomes insensitive to the insulin fails to bring GLUT 4 to the surface of the cell
answer
type 2 diabetes
question
dihydroxyacetone phosphate intermediate of glycolysis used in hepatic and adipose tissue for triglyceride synthesis formed from fructose 1,6 phosphate isomerized into glycerol 3 phosphate glycerol 3 phosphate can then be converted into glycerol glycerol then acts as a triglyceride backbone
answer
DHAP
question
all cells can do this cytoplasmic pathway - in cytoplasm converts *1 glucose to 2 pyruvates* gives the intermediated for other pathways releases energy through two substrate level phosphorylations and one oxidative reaction - uses up 2 ATP - x2(makes 2 ATP) -x2 (makes 1 NADH) overall = 1 glucose --> 2 pyruvates 2 ATP 2NADH important enzymes hexokinase/ glucokinase PFK1/ PFK2 aldoase glyceraldehyde 3 phosphate dehydrogenase phosphoglycerate kinase pyruvate kinase in liver - part of the process that excess glucose is converted to fatty acids for storage (due to PFK2 allows for PFK1 to continue even when have enough ATP) can feed into aerobic respiration if have mitochondria or oxygen is present because NADH (carrier product of this) can feed into it if lacks mitochondria or oxygen (RBC, exercising skeletal muscle), can occur anaerobically and go into fermentation - some energy may be lost glucose, galactose and fructose can all lead into this pathway
answer
glycolysis
question
are needed for: citric acid cycle electron transport chain oxidative phosphorylation fatty acid metabolism,/ Beta Oxidation
answer
mitochondria
question
irreversible enzyme in glycolysis found in a lot of cells is inhibited by *glucose 6 phosphate* - its product (product inhibition) attaches a phosphate group from ATP to glucose this prevents the glucose from leaving the cell once it has entered can also slowly phosphorylate fructose glucose --> glucose 6 phosphate low KM - reaches maximum velocity when concentrations of glucose are low
answer
hexokinase
question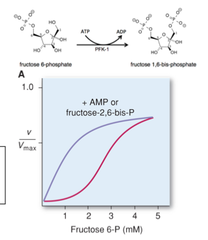
phosphofructokinase 1 *RATE LIMITING ENZYME OF GLYCOLYSIS* -main point of control for glycolysis irreversible enzyme phosphorylates fructose 6 phosphate at the one position fructose 6 phosphate --> fructose 1,6 phosphate inhibited by: turn off glycolysis when there is enough ATP/ energy ATP CITRATE glucagon (by PFK2) activated by: turn on glycolysis when need more energy AMP insulin (by PFK2) (insulin activated PFK2, which creates fructose 2,6 phosphate - fructose 2,6 phosphate activated this enzyme!)

answer
PFK1
question
phosphofrunctokinase 2 activated by *insulin* inhibited by *glucagon* fructose 6 phosphate --> fructose 2,6 phosphate fructose 2,6 phosphate activates PFK1 found mostly in the liver by activating this (due to insulin) it allows PFK1 to be activated and continued even though there is enough ATP - allows the production of glycogen, fatty acids and storage molecules to be made - allows storage, and not just ATP to be burned
answer
PFK2
question
enzyme in glycolysis does an oxidation and an addition of inorganic phosphate to glyceraldehyde 3 phosphate to 1,3 bisphosphoglycerate (1,3 BPG) also reduces NAD+ to NADH

answer
glyceraldehyde 3 phosphate dehydrogenase
question
enzyme in glycolysis transfers the high energy phosphate group of 1,3 bisphosphoglycerate to ADP - thus forms ATP
answer
3 phosphoglycerate kinase
question
ADP is directly phosphorolated to ATP by a high energy intermediate not dependent on oxygen (different than oxidative phosphorylation) - only means of ATP in an anaerobic organism done by 3 phosphoglycerate kinase
answer
substrate level phosphorylation
question
irreversible enzyme in glycolysis last enzyme does substrate level phosphorylation takes PEP and transfers its phosphate group to an ADP to create ATP and pyruvate activated by fructose 1,6 phosphate (made by PFK1) - feed forward reaction
answer
pyruvate kinase
question
phosphoenolpyruvate high energy substrate of glycolysis used to generate ATP through substrate level phosphorylation pyruvate kinase takes a phosphate group from this and transfers it to ATP and creates pyruvate in the last step of glycolysis
answer
PEP
question
product of an earlier reaction stimulates/ prepares a later reaction ex. PFK 1 makes fructose 1,6 phosphate which stimulates pyruvate kinase (another enzyme) necessary to make pyruvate and finish glycolysis ex. glucose 6 phosphate stimulates glycogen synthase in glycogenesis
answer
feed forward reaction
question
will occur after glycolysis if no oxygen is present main goal = REPLENISH NAD+ key enzyme = *lactate dehydrogenase* pyruvate is converted to lactate while NADH is oxidized to NAD+ this allows NAD+ to go back to glycolysis and continue glycolysis when oxygen levels are low (strenuous exercise, heart attack, stroke) lactate is produced in yeast cells= pyruvate is converted to ethanol and carbon dioxide
answer
fermentation
question
RATE LIMITING ENZYME in fermentation oxidizes NADH to NAD+ while pyruvate is converted to lactate
answer
lactate dehydrogenase
question
oxidation reaction
answer
NADH to NAD+
question
reduction reaction
answer
NAD+ to NADH
question
1,3 bisphosphoglycerate high energy substrate in glycolysis made by glyceraldehyde 3 phosphate dehydrogenase from glyceraldehyde 3 phosphate used to generate ATP through substrate level phosphorylation
answer
1,3 BPG
question
glucokinase/hexokinase PFK1 pyruvate kinase different enzymes must be used in gluconeogenesis
answer
irreversible enzymes of glycolysis
question
no mitochondria! have bisphoglycerate mutase
answer
glycolysis in erythrocytes
question
enzymes that move a functional group from one place in the molecule to another
answer
mutases
question
found in RBC converts 1,3 BPG to 2,3 BPG
answer
bisphoglycerate mutase
question
2,3, bisphosphoglycerate converted from 1,3 BPG by bisphoglycerate mutase in red blood cells phosphate got moved to the two position binds to the Beta chain of hemoglobin which decreases the affinity for oxygen creates a rightward shift in the oxygen dissociation curve *allows an unloading of oxygen in tissues but still allows for 100% saturation in the lungs*
answer
2,3 BPG
question
at low oxygen... increased respiration - trying to get more oxygen increased affinity for oxygen increased rate of glycolysis increased 2,3,BPG - allows an unloading of oxygen in tissues but still allows for 100% saturation in the lungs normalized oxygen affinity for hemoglobin restored by the increased level of 2,3 BPG increase amount of hemoglobin
answer
adaption to high altitudes/ low oxygen
question
high 2,3 BPG low PH high H+ high CO2 low O2 all of these occur during exercise
answer
right shift of the oxygen dissociation curve
question
lactose is broken down into galactose and glucose by lactase galactose is phosphorylated by *galactokinase* to trap it inside the tissues/ cells galactose 1 phosphate is converted into glucose 1 phosphate by *galactose 1 phosphate uridyltransferase*
answer
galactose metabolism
question
breaks down lactose into galactose and glucose brush boarder enzyme in the duodenum
answer
lactase
question
traps galactose inside the cell by phosphorylating it galactose --> galactose 1 phosphate
answer
galactokinase
question
converts galactose 1 phosphate to glucose 1 phosphate epimerase
answer
glactose 1 phosphate uridyltransferase
question
enzymes that catalyze the conversion of one sugar epimer to another
answer
epimerase
question
liver phosphorylated fructose to trap it by *fructokinase* making fructose 1 phosphate *adolase B* than cleave fructose 1 phosphate into DHAP and glyceraldehyde to be used in glycolysis *since these products are down stream of the rate limiting enzyme of glycolysis (PFK1) - high fructose drinks gives the person a lot of energy/ storage*
answer
fructose metabolism
question
found in honey and fruit comes from the break down of sucrose by sucrase
answer
fructose
question
brush boarder enzyme that breaks down sucrose into fructose and glucose
answer
sucrase
question
found in the liver phosphorylates fructose into fructose 1 phosphate this traps fructose inside the cell
answer
fructokinase
question
pyruvate dehydrogenase part of the pyruvate dehydrogenase complex irreversible (cannot get pyruvate or glucose from acetyl CoA) inhibited by its product - acetyl CoA needs: thiamine pyrophosphate lipoic acid CoA FAD NAD+ oxidizes pyruvate creating CO2 the remaining two carbon molecule (use to be pyruvate) is connected to TPP) activated by insulin in the liver - high insulin levels signal that the person is in a fed state so should not burn glucose for energy but go into Beta oxidation for fatty acid synthesis in nervous system this is not effected by hormones takes pyruvate (after it enters the mitochondria) and converts it to acetyl CoA
answer
PDH
question
needed to start the citric acid cycle when ATP is needed needed to for fatty acid synthesis when ATP is already present
answer
acetyl CoA
question
1. conversion to acetyl CoA by PDH 2. conversion to lactate by lactate dehydrogenase 3. conversion to oxaloacetate by pyruvate carboxylase when there is a build up of acetyl CoA, this inhibits PDH and so the CoA goes to Beta Oxidation also pyruvate is no longer converted into acetyl CoA but goes to make oxaloacetate
answer
fates of pyruvate
question
converts pyruvate to oxaloacetate by adding a carboxyl group to it mitochondria enzyme activated acetyl CoA from Beta Oxidation (the acetyl CoA is from fatty acids) fatty acids must be burned in order to provide enough energy to stop the citric acid cycle and produce massive amounts of oxaloacetate that can eventually lead into glucose production for the rest of the body happens when acetyl CoA is high (inhibits PDH but activates this) part of the gluconeogenesis pathway
answer
pyruvate carboxylase
question
vitamin B1 cofactor needed in the PDH complex can cause Beriberi and Wernicke - Korsafoff syndrome
answer
deficiency in thiamine
question
due to a deficiency in thiamine has congestive heart failure and nerve damage
answer
beriberi
question
due to a deficiency in thiamine difficulty walking uncoordinated eye movement confusion memory disturbances
answer
Wernicke - Korsafoff syndrome
question
branched polymer of glucose storage form of glucose synthesis and degradation - occurs in the liver and skeletal muscle stores in the cytoplasm as granules - each granule has a protein core with poly glucose chains coming off of it in the liver the break down of it is to maintain a constant level of glucose in the blood - mobilized during meals to prevent low blood sugar in the muscle the break down of it is to provide glucose to the muscle during exercise
answer
glycogen
question
long alpha linked chains of glucose way plants store glucose
answer
starch
question
synthesis of glycogen granules 1. the making of glycogenin 2. glucose 6 phosphate is converted to glucose 1 phosphate 3. glucose 1 phosphate is activated when it is coupled to UTP -this creates glucose 1 phosphate-UDP and PPi (pyrophasophate) 4. *Glycogen synthase* (Rate limiting) - makes the alpha 1,4 glycosidic bonds 5. Branching enzyme makes the 1,6 linked branches when it is coupled to UDP glycogen synthase takes glucose 1 phosphate and UDP onto glycogen
answer
glycogenesis
question
core protein found in the glycogen protein start of glycogenesis
answer
glycogenin
question
PPi
answer
pyrophosphate
question
RATE LIMITING enzyme of glycogenesis responsible for making the alpha 1,4 glycosidic bonds as the granule grows stimulated by insulin and the reactants (glucose 6 phosphate) --> example of feed forward inhibited by epinephrine and glucagon (through a protein cascade that phosphorylates it and inactivates it)
answer
glycogen synthase
question
makes the alpha 1,6 branches found in glycogen hydrolyses the alpha 1,4 glycosidic linkage and than attaches those few chained glucose molecules to another spot of the chain and forms a alpha 1,6 bond to finish the branch
answer
branching enzymes
question
break down of glycogen rate limiting enzyme: glycogen phosphorylase glycogen phosphorylase breaks down the alpha 1,4 glycosidic bonds debranching enzyme the residue that contains the alpha 1,6 bond is the only free glucose made directly in this pathway the other glucose comes from glucose 1 phosphate to glucose 6 phosphate by a mutase glucose 6 phosphate is transfered to glucose by glucose 6 phosphatase
answer
glycogenolysis
question
rate limiting enzyme in glycogenolysis breaks the alpha 1,4 glycosidic bonds of glycogen releases glucose-1-phosphate into the circulation cannot break alpha 1,6 branch points and stops when it gets to these branch points activated by glucagon in the liver - so it can provide glucose to the rest of the body activated by epinephrine and AMP in the muscle - because those are indicators that the muscle is active and requires glucose inhibited by ATP
answer
glycogen phosphorylase
question
breaks bonds using an inorganic phosphate instead of water (hydrolase) glucose 1 phosphate --> glucose 6 phosphate
answer
phosphorylase
question
enzyme in glycogenolysis two enzyme complex takes the alpha 1,6 branching points that have been exposed due to glycogen phosphorylase moves the whole branch (besides the residue that is making the alpha 1,6 linkage) to the terminal end of the chain making a new alpha 1,4 glycosidic bond (alpha 1,4 transferase) the other removes the glucose monomer that was actually present at the branch point (alpha 1,6 glucosidase) hydrolyses the alpha 1,6 bond
answer
debranching enzyme
question
slightly different versions of the same protein
answer
isoforms
question
characterized by the accumulation or lack of glycogen in one or more tissues due to isoforms of glycogen enzymes most common is Von Gierke's disease
answer
glycogen storage disease
question
most common glycogen storage disease defect in glucose 6 phosphatase leads to periods of very low blood sugar between meals needs continuous feeding to maintain blood sugar levels glucose 6 phosphate builds up in the cell and cannot be broken down into glucose - so the liver enlarges and is damaged over time
answer
Von Gierke's disease
question
enzyme that converts glucose 6 phosphate to glucose in glycogenolysis last step in gluconeogenesis defected in Von Gierke's disease
answer
glucose 6 phosphatase
question
liver does this in times of fasting also can be done by the kidneys after fasting for 12 hours, blood glucose drops drastically which is when this increases. After 24 hours of this it is the sole source of glucose in the blood activated by: - act to raise blood sugar glucagon epinephrine inhibited by: insulin - acts to lower blood sugar important substrates: 1. glycerol 3 phosphate - from stores fats/triacylglercols, adipose tissue *glycerol 3 phosphate is converted to DHAP by glycerol 3 phosphate dehydrogenase* 2. lactate - from anaerobic glycolysis *lactate is converted into pyruvate by lactate dehydrogenase* 3. glucogenic amino acids - from muscle proteins *alanine is converted to pyruvate by alanine aminotranferase* once has pyruvate basically does the exact reversal of glycolysis besides at the irrveersable steps! uses these enzymes: pyruvate carboxylase PEPCK Fructose 1,6 Bisphosphatase - RATE LIMITING ENZYME glucose 6 phosphatase pyruvate carboxylase and PEPCK are two enzymes used to convert pyruvate back into pyruvate in anaerobic - pyruvate converted to lactate to regenerate NAD+m lactate is acidic so goes to the liver and converted back into pyruvate via this mechanism = Cori cycle lactate is converted to glucose in liver!
answer
gluconeogenesis
question
lysine and leucine are not included in this these amino acids can be converted into intermediates and go into gluconeogenesis
answer
glucogenic amino acids
question
amino acids that can be converted into acetyl CoA and ketone bodies lysine, leucine, isoleucine, phenylalanine, theronine, trypophan, tryosine
answer
ketogenic amino acids
question
glycogenolysis gluconeogenesis galactose and fructose not fatty acids (they get broken down into acetyl CoA which cannot go back into pyruvate/ glucose) - unless create *propinoly-CoA* from an odd number of fatty acids
answer
sources of glucose
question
enzyme in gluconeogenesis found in the cytoplasm started by glucagon and cortisol converts oxaloacetate to PEP needs GTP
answer
PEPCK
question
enzyme in gluconeogenesis found in cytoplasm RATE LIMITING ENZYME reverses the action of PFK1 fructose 1,6 phosphate --> fructose 6 phosphate activated by ATP inhibited by AMP and fructose 2,6 biphosphate
answer
Fructose 1,6 bisphosphatase
question
substrate that comes from PK2 in glycolysis controls both gluconeogenesis and glycolysis in the liver PK2 activated by insulin and deactivated by glucagon glucagon inhibit PK2 and lower this which will stimulate gluconeogenesis insulin will promote PK2, increasing this and inhibiting gluconeogenesis
answer
fructose 2,6 phosphate
question
found in the lumen of the ER in liver cells enzyme in gluconeogenesis converts glucose 6 phosphate to glucose glucose 6 phosphate is transported to the ER and the free glucose comes from the ER and into the cytoplasm where it can leave through the GLUT transporters not found in skeletal muscle - this means that the glycogen cannot serve as a source of blood glucose and only use it for the muscle itself reverse of hexokinase/ glucokinase
answer
glucose 6 phosphatase
question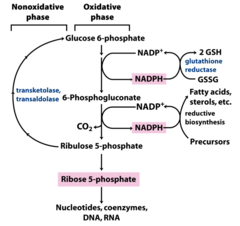
PPP also known as the *hexose monophosphate (MPH) shunt occurs in the cytoplasm of all cells functions: 1. production of NADPH 2. gives a source of ribose 5 phosphate for nucleotide synthesis 1. G6PD = rate limiting enzyme glucose 6 phosphate NADP+ --> ribulose 5 phosphate + NADPH 2. equilibrium pool with other sugars like ribose 5 phosphate (backbone for nucleic acids) for nucleotide synthesis fructose 6 phosphate glyceraldehyde 3 phosphate sugars produced - can feed back into glycolysis transketolase and transaldolase - enzymes that can makes form pentoses with out going through G6PD

answer
Pentose Phosphate Pathway
question
glucose 6 phosphate dehydrogenase rate limiting enzyme of the pentose phosphate pathway converts glucose 6 phosphate to ribulose 5 phosphate also by making NADPH activated by insulin - the abundance of sugar entering the cell under insulin stimulation will be shunted into both glycolysis and aerobic respiration as well as all the storage pathways (fatty acid synthesis glycogenesis, PPP) also activated by reactants = NADP+ inhibited by the product = NADPH
answer
G6PD
question
acts as a electron donor in biological reactions reducing agents - helps other molecules be reduced while it is oxidized in the process 1. need it for the biosynthesis of fatty acids and cholesterol 2. is involved in the bleach production of white blood cells - contributing to the bactericidal activity 3. maintains a reduced supply of glutathione - which protects against reactive oxygen species - bodies natural antioxidant 4. protects cells against free radicals caused by peroxides (by product of aerobic respiration) - free radicals can attack against lipids - those found in the membrane, if attack can weaken the membrane and cause the cell to lyse
answer
NADPH
question
high energy electron acceptor for a number of biological reactions oxidizing agent - helps the other molecule be oxidized while itself is reduced in the process
answer
NAD+
question
high energy electron acceptor for a number of biological reactions oxidizing agent - helps the other molecule be oxidized while itself is reduced in the process
answer
NADP+
question
acts as a electron donor in biological reactions reducing agents - helps other molecules be reduced while it is oxidized in the process
answer
NADH
question
reducing agent that can help reverse radical formation before damage is done to the cell NADPH makes a reduced supply of this
answer
glutathione
question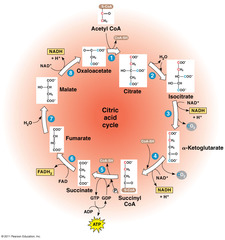
TCA Cycle or Krebs Cycle occurs in the mitochondria matrix oxidation of acetyl CoA to CO2 and H2O also produces high energy electron carrying molecules (NADH and FADH2) will not occur anaerobically - because NADH and FADH2 will accumulate if oxygen in not available for electron transport chain Actyl CoA +3NAD+ + FAD + GDP + P + 2H2O --> 2CO2 + COASH + 3 NADH + 3H+ +FADH2 + GTP key reactions: Step 1: Citrate formation Step 2: Citrate isomerized to Isocitrate Step 3: alpha ketoglutarate and CO2 formation Step 4: Succinyl CoA and CO2 formation Step 5: Succinate Formation Step 6: Fumarate Formation Step 7: Malate Formation Step 8: Oxaloacetate Formed Anew Please, Can I Keep Selling Seashells For Money Officer? Pyruvate, Citrate, Isocitrate, (oxalosuccinate), alpha Ketoglutarate, Succinyl- CoA, Succinate, Fumarate, Malate, Oxaloacetate

answer
Citric Acid Cycle
question
1. from glucose glucose undergoes glycolysis to make pyruvate, which enters the mitochondria via active transport - then it is oxidized and decarboxylated by PDH complex (located in the matrix of the mitochondria) pyruvate --> acetyl CoA + CO2 exergonic = -G inhibited by NADH and acetyl CoA 2. from fatty acids - Beta oxidation 3. from ketone bodies ketone bodies to acetyl CoA (usually acetyl CoA is used to make ketone bodies when PDH is inhibited) 4. from alcohol enzymes: *alcohol dehydrogenase and acetaldehyde dehydrogenase* they convert alcohol to acetyl CoA this reaction is accompanied by NADH build up - inhibits the Citric Acid Cycle *acetyl CoA formed through this process is used primarily to synthesize fatty acids* 5. from amino acids - amino acid catabolism the amino acid must lose their amino group via transamination the carbon skeletons then form the ketone bodies - ketogenic amino acids
answer
methods of forming acetyl CoA
question
made up of five enzymes 1. PDH 2. dihydrolipoyl transacetylase 3. dihydrolipoyl dehydrogenase 4. pyruvate dehydrogenase kinase 5. pyruvate dehydrogenase phosphatase PDH (pyruvate + TPP--> acyl-TPP--PDH + CO2) -Mg2+ is also needed for this pyruvate + COASH + NAD+ --> acetyl CoA + NADH + CO2 + H+
answer
pyruvate dehydrogenase complex
question
written as CoA-SH CoA is a thiol (contains a SH group) when acetyl CoA forms it does so by a covalent attachment of the acetyl group to the SH group - resulting an a thioester instead of a OR thioester contains more energy than a normal ester (OR) when a thioester undergoes hydrolysis - a significant amount of energy is released - this energy will help push the other reactions forward
answer
Coenzyme A
question
vitamin B1 holds the two carbon molecule that use to be pyruvate is a coenzyme that is held to PDH by noncovalent interactions
answer
TPP
question
enzyme in the PDH complex takes the two carbon molecule attached to TPP and transfers it to lipoic acid acetyl group is now attached to the lipoic acid via a thioester linkage COASH then comes in making acetyl CoA and reduced lipoic acid
answer
dihydrolipoyl transacetylase
question
dihydrolipoyl transacetylase transfres the two carbon molecule from TPP to this it is an coenzyme covalently attached to the PDH complex has disulfide bonds that act as oxidizing agents - taking electrons creates the acetyl group
answer
lipoic acid
question
enzyme in the PDH complex used FAD to re oxidize lipioc acid so it is ready to be reduced once more lip -SH-SH +FAD+ --> LipS-S + FADH2 NAD+ oxidizes FADH2 to FAD+ and NADH
answer
dihydrolipoyl dehydrogenase
question
1. activation in the intermembrane space = fatty acyl- CoA 2. fatty acyl Co-A is then transported into the intermembrane space 3. fatty acyl CoA cannot cross the inner mitochondria membrane - fatty acid is transferred to a carnitine via transesterification reaction 4. carnitine with fatty acid can go through the inner mitochondria membrane 5. once in the matrix the fatty acyl group is transferred to another CoASH in another transesterification reaction now the beta oxidation can occur - taking two carbon fragments from the carboxyl end
answer
beta oxidation
question
happens in the intermembrane space first step of beta oxidation thioester bonds forms between the carboxyl group of free fatty acids and Co-ASH = fatty acyl CoA
answer
activation
question
first step of the TCA cycle acetyl CoA and oxacloacetate go through a *condensation reaction* reaction enzyme: *citrate synthase* (synthases - enzymes that form new covalent bonds without needing significant energy) - helps move the reaction forward *checkpoint for TCA cycle* COASH is a product acetyl COA + oxaloacetate --> COASH + citrate
answer
Step 1: Citrate formation
question
2nd step of the TCA cycle citrate is isomerized into one of four isomers of isocitrate citrate binds to three points on *aconitase* water is lost from citrate = creating a double bond = cis- aconitase water is added back to form isocitrate aconitase is a metaloprotein - requires Fe2+ and a hydroxyl group needed because puts the carboxyl group in the beta place for beta decarbozylation
answer
Step 2: Citrate isomerized to Isocitrate
question
3rd step of the TCA cycle isocitrate is oxidized to oxalosuccinate by *isocitrate dehydrogenase* isocitrate dehydrogenase = RATE LIMITING ENZYME oxalocussicnate is DECARBOXYLATED to make alpha ketoglutarate and CO2 *first CO2 lost and first NADH is made*
answer
Step 3: alpha ketoglutarate and CO2 formation
question
4th step of the TCA cycle reactions are carried out by *alpha ketoglutarate dehydrogenase complex*- have similar cofactors and coenzymes as the PDH complex alpha ketoglutarate and COASH come together, the form CO2 and succinyl CoA second CO2 is lost 2nd NADH is formed here
answer
Step 4: Succinyl CoA and CO2 formation
question
fifth step of the TCA cycle *succinyl CoA synthetase* hydrolysis the thioester bond in succinyl CoA to make succinate and COASH the hydrolysis of this thioester bond creates sufficient energy to phosphorylate GDP to GTP
answer
Step 5: Succinate Formation
question
sixth step of the TCA cycle only step that does NOT take place in the matrix - it takes place in the inner membrane succinate undergoes oxidation to form fumarate by *succinate dehydrogenase* - integral protein on the inner membrane and attached to FAD as succinate is oxidized, FAD is reduced to FADH2 FADH2 than passes the electrons to eventually produce 1.5 ATP
answer
Step 6: Fumarate Formation
question
seventh step of the TCA cycle enzyme *fumerase* hydrolyzes (input of water) the alkene bond in fumerate this creates malate L malate is the only one that forms
answer
Step 7: Malate Formation
question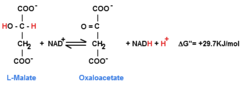
eighth step of the TCA cycle *malate dehydrogenase* oxidizes malate to oxaloacetate NAD+ is reduced to NADH

answer
Step 8: Oxaloacetate Formed Anew
question
rate limiting enzyme of the TCA cycle check point of TCA cycle makes isocitrate oxidized into oxalosuccinate, which can spontaneously go through Beta decarboxylation inhibited by ATP and NADH ADP and NAD+ activate the enzyme by increasing its affinity for substrates
answer
isocitrate dehydrogenase
question
checkpoint for TCA cycle needs TPP, lipoic acid, and Mg2+ converts alpha ketoglutarate to succinyl CoA inhibited by succinyl CoA, ATP, NADH stimulated by ADP and calcium ions
answer
alpha ketoglutarate dehydrogenase complex
question
create new covalent bonds WITH energy input ex. succinyl CoA synthetase
answer
synthetases
question
create new covalent bonds WITHOUT energy input ex. citrate synthase
answer
synthases
question
enzyme in the citric acid cycle that synthesizes the transfer of a phosphate from GTP to ADP creating ATP
answer
nucleosidediphosphate kinase
question
oxidizes succinate to fumarate considered a flavoprotein - because it is covenantally connected to FAD integral protein on the inner membrane as succinate is oxidized, FAD is reduced to FADH2 FADH2 than passes the electrons to eventually produce 1.5 ATP
answer
succinate dehydrogenase
question
FOR EVERY Pyruvate glycolysis 1 ATP 1 NADH --> 2.5 ATP 3.5 PDH complex 1 NADH --> 2.5 ATP 2.5 TCA cycle 3 NADH --> 7.5 ATP 1 FADH2 --> 1.5 ATP 1 GTP --> 1 ATP 10 16 ATP per pyruvate 32 ATP per glucose
answer
Net ATP
question
regulated by pyruvate dehydrogenase kinase and pyruvate dehydrogenase phosphatase
answer
pyruvate dehydrogenase complex regulation
question
inactivates PDH by phosphorylating it by phosphorylating it when ATP levels rise, it can no longer make acetyl CoA
answer
pyruvate dehydrogenase kinase
question
reactivates PDH in response to high amounts of ADP removes a phosphate group from PDH
answer
pyruvate dehydrogenase phosphatase
question
three checkpoints 1. Citrate synthase ATP, NADH, citrate, succinyl CoA function as allosteric inhibitors of this enzyme 2. isocitrate dehydrogenase inhibited by ATP and NADH ADP and NAD+ activate the enzyme by increasing its affinity for substrates 3. alpha ketoglutarate dehydrogenase complex inhibited by succinyl CoA, ATP, NADH stimulated by ADP and calcium ions
answer
control points of the citric acid cycle
question
*the proton gradient - not the flow of electrons - produces the ATP* generate ATP using a proton motive force generates ATP through ADP phosphorylation NADH and FADH2 transfer their electrons to carrier proteins in the inner mitochondria membrane the electrons are given to oxygen in the form of hydride ions (H-) and water is formed as the electrons are being transported to oxygen protons are being transported into the intermembrane space of the mitochondria = creating a greater concentration gradient of hydrogen ions that can be used for ATP production formation of ATP = endergonic electron transport = exergonic pathway physical property that determines the direction of electron flow = reduction potential molecule with the higher reduction potential will be the one that gets reduced and the other will be oxidized no more than a series of oxidations and reductions that occur high reduction potential of oxygen makes it a great final acceptor COMPLEX 1 COMPLEX 2 COMPLEX 3 COMPLEX 4
answer
electron transport chain
question
NADH CoQ oxidoreducatse oxidation of NADH transfer of electrons/ H- to the coenzyme Q (CoQ) has two major subunits 1. iron sulfur cluster 2. flavoprotein with FMN (flavin mononucleotide) coenzyme FMN has similar structure to FAD as NADH gets oxidized to NAD+ FMN goes to FMNH2 (reduced) FMNH2 gets oxidized to FMN iron sulfur cluster gets reduced reduced iron sulfer subunit gives electrons to CoQ/ ubiquinone CoQ --> CoQH2 *four protons are moved to the intermembrane space* net: NADH + H+ + CoQ --> NAD+ + CoQH2
answer
COMPLEX 1 of ETC
question
ubiquinone
answer
coenzyme q
question
succinate CoQ oxidoreductase receives electrons from succinate as it is formed into fumerate FAD is covalently bonded to this and once succinate is oxidized into fumerate, FADH2 is formed iron sulfur protein oxidizes FADH2 back to FAD the iron sulfur protein transfers the electrons to coenzyme Q *no hydrogen pumping occurs here* net: succinate + CoQ + 2H --> fumerate +CoQH2
answer
COMPLEX 2 of ETC
question
CoQH2 cytochrome c oxidoreductase also called *cytochrome reductase* this complexes allows the transfer of electrons from coenzyme Q to cytochrome c the steps involve the oxidation and reduction of cytochromes CoQH2 + 2 cytochrome c with Fe3+ --> CoQ + 2 cytochrome c with Fe2+ + 2H only on electron can be transferred to iron, which is why you need two cytochromes *q cycle is done here* - pumping *four protons*
answer
COMPLEX 3 of ETC
question
proteins with heme proteins in which iron is reduced to Fe2+ and then deoxidized to Fe3+
answer
cytochromes
question
two electrons are shuttled from a molecule of ubiquinol (CoQH2) near the intermembrane space to a ubiquinone (CoQ) need the matrix another two molecules are attached to heme groups - reducing two cytochrome c's (group with iron and sulfur helps this) *four protons are pumped into the intermembrane space in this process* - continues to increase the gradient of the proton motive force across the inner mitochondria membrane
answer
Q cycle
question
cytochrome c oxidase facilitates the final transfer of electrons from cytochrome c to oxygen (final electron acceptor) has subunits of cytochrome a, cytochrome a3 and Cu2+ ions cytochrome oxidase (a and a3) gets oxidized as oxygen gets reduced to water final location where proton pumping occurs - two protons are moved across the membrane net: 4 cytochrome c with Fe2+ + 4H+ + O2 --> 4 cytochrome c with Fe3+ + 2H2O
answer
COMPLEX 4 of ETC
question
cytochromes a and a3 in complex 4
answer
cytochrome oxidase
question
electrostatic gradient that is based on the protons a proton gradient is formed as electrons are passed through the ETC as H+ increase in the intermembrane space = 1. pH drops 2. the voltage difference between the intermembrane space and the matrix increases due to proton pumping this force is capable of storing energy
answer
proton motive force
question
gradient that has both chemical and electrostatic properties stores energy
answer
electrochemical gradient
question
harness the energy which is created due to the proton motive force and forms ATP from ADP and an inorganic phosphate
answer
ATP synthase
question
glucose yields 30-32 ATP cytosolic NADH through glycolysis cannot cross the mitochondria matrix - because it cannot contribute its electrons to the ETC directly it has to find other ways into the matrix the ways of transportation into the matrix will depend on if 30 or 32 ATP's are made two different shuttle mechanisms: 1. *glycerol 3-phosphate shuttle* 2. *Malate aspartate shuttle*
answer
Variability of glycolysis
question
DHAP is transferred to glycerol 3 phosphate by an isoform of *glycerol 3 phosphate dehydrogenase enzyme* which is *FAD dependent* FAD is the oxidizing agent and as glycerol 3 phosphate is made FAD goes to FADH2 FADH2 than goes into the ETC yielding 1.5 ATP
answer
glycerol 3-phosphate shuttle
question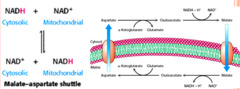
cytosolic oxaloacetate cannot pass through the inner membrane so oxaloacetate is transferred into malate through *malate dehydrogenase* and the oxidation of NADH to NAD+ malate crosses the inner membrane and then reverse the reaction to form NADH NADH can now go into the ETC making 2.5 ATP

answer
Malate aspartate shuttle
question
inhibitor of cytochrome a and a3 inhibitor of complex 4 cyanide ion is able to attach to the iron group and prevent the transfer of electrons cyanide poisoning
answer
cyanide
question
done by ATP synthase
answer
oxidation phosphorylation
question
ion channel in ATP synthase protons go through this along their gradient back into the mitochondrial matrix
answer
F0
question
describes a direct relationship between the proton gradient and ATP synthase allows the chemical energy of the gradient to be harnessed as means of phosphorylating ADP and creating ATP mechanism states that the ETC generates a high concentration of protons in the intermediate space which then flow through F0 ion channel of ATP syntheses back into the matrix - as this happens f1 utilizes the energy released from the electrochemical gradient to phosphorylate ADP
answer
chemiosmotic coupling
question
utilizes the energy released from the electrochemical gradient to phosphorylate ADP to ATP
answer
F1 portion of ATP synthase
question
suggests that the relationship between the proton gradient and the ATP synthase is indirect in this mechanism - the F1 portion of ATP synthase is like a turbine, spinning within a stationary compartment to facilitate the harnessing of gradient energy for chemical bonds
answer
conformational coupling
question
rates of oxidation phosphorylation and citric acid cycle are very closely linked - the TCA cycle gives the reactants for the ETC O2 and ADP are key regulators O2 limited rate of oxidation phosphorylation decreases concentration of NADH and FADH2 increase - inhibiting the TCA cycle enough O2 - dependent of the availability of ADP ADP and ATP are reciprocally related a lot of ADP - not much ATP a lot of ATP - not much ADP a lot of ADP - need for ATP synthesis ADP activated isocitrate dehydrogenase - increasing the rate of the TCA cycle and thus increasing the production of NADH and FADH2
answer
respiratory control
question
compounds that prevent ATP synthesis without effecting the ETC and thus decreasing the efficiency of the ETC/oxidative phosphorylation pathway ADP builds up ATP synthesis decreases body sense lack of energy production - increases O2 production and NADH oxidation energy from the electrons is released as heat - fever that rises from toxic levels of salicylates (asprin)
answer
uncouplers
question
consists mainly of tricylgylcerols rest of it is cholesterol, cholestryl esters, phospholipids and free fatty acids
answer
dietary fat
question
minimal in mouth and stomach go to the small intestine essentially intact emulsification occurs right before the entry to the duodenum - water and fat is now mixed which increases the surface area of the lipid and permits greater enzymatic interactions emulsification is aided by bile (bile salts, pigments, cholesterol) which is secreted by liver and stored in the gallbladder pancreas secreted pancreatic lipase, colipase, cholesterol esterase into the small intestine now lipid components are *2 monoacylglycerol, free fatty acids and cholesterol* this forms micelles end of the ileum bile salts are reabsorbed and recycled - any fat that remains in the intestine will pass into the colon and end up in poo
answer
digestion of fats
question
the free fatty acids, 2 monoacyclglycerol, cholesterol with bile salts all form into micelles
answer
micelle formation
question
clusters of amphipathic lipids soluble in aqueous environments of the intestinal lumen water soluble spheres with a lipid soluble interior they will diffuse to the brush boarder of the intestine mucosal cells where they are then absorbed
answer
micelle
question
micelles will diffuse to the brush boarder of the intestine mucosal cells where they are then absorbed they are then re-esteried to form triglycerides ad cholestryl esters and are packed with the fat soluble vitamins into *chylomicrons* chylomicrons lave the intestine via lacteals they re enter the blood stream via tha thoracic duct
answer
absorption of micelle
question
long lymphatic vessel that empties into the left subclavian vein at the base of the neck fat re enters the blood here
answer
thoracic duct
question
vessels of the lymphatic system fat leaves the intestine and goes into these
answer
lacteals
question
hormone sensitive lipase hydrolyses triglycerides triglycerides --> free fatty acids + glycerol fall of insulin activates this epinephrine and cortisol activate this glycerol goes to the liver and is used for glycolysis and gluconeogenesis
answer
HSL
question
lipoprotein lipase enzyme that releases the free fatty acids and triglycerides in the lipoproteins necessary for the metabolism of chylomicrons and VLDL
answer
LPL
question
carrier protein that transports free fatty acids through the blood
answer
albumin
question
proteins with lipid prosthetic groups transports cholesterol and triglycerides they are aggregates of apolipoproteins and lipids names according to density of proteins 1. Chylomicron 2. VLDL 3. IDL 4. LDL 5. HDL
answer
lipoproteins
question
type of lipoprotein least dense with protein highest fat to protein ratio highly soluble in lymphatic and blood fluid transport dietary triglycerides, cholesterol, and cholesteryl esters from intestine to tissues assembly of these occurs in the intestinal lining
answer
chylomicron
question
type of lipoprotein a little bit more dense with proteins then chylomicrons transports triglycerides and fatty acids from the liver to tissues assembled in the liver cells contain fatty acids that are synthesized from excess glucose or retrieved from chylomicron remnants
answer
VLDL
question
type of lipoprotein intermediate protein density slightly more dense with protein than VLDL known as this or VLDL remnant when the triglyceride is removed from the VLDL some of them are reabsorbed by the liver by apolipoproteins on the exterior some are further processed in the blood stream picks up cholesteryl esters from HDL to become LDL picked up by the liver *exists as a transition molecule between triglyceride transport (association with chylomicrons and VLDL) and cholesterol transport (associated with LDL and HDL)*
answer
IDL
question
type of lipoprotein low density of proteins delivers cholesterol into cells cholesterol in the blood is associated with this tissues might need the cholesterol for their membranes, steroidogenesis, to make bile s
answer
LDL
question
type of lipoprotein high density of proteins picks up cholesterol accumulating in the blood vessels delivers cholesterol to liver and steriodogenic tissues transfers apolioproteins to other lipoproteins synthesized in the liver and intestines contains apolipoproteins - used for cholesterol recovery
answer
HDL
question
form a protein component of the lipoproteins they are receptor molecules and involved in signaling apoA-I apoB-48 apoB-100 apoC-II apoE
answer
apolipoproteins
question
apolipoprotein activates LCAT
answer
apoA-I
question
apolipoprotein mediates chylomicron secretion
answer
apoB-48
question
apolipoprotein permits uptake of LDL by the liver
answer
apoB-100
question
apolipoprotein actiavtes lipoprotein lipase
answer
apoC-II
question
apolipoprotein permits uptake of chylomicron remnants and VLDL by the liver
answer
apoE
question
lecithin- cholesterol acyltransferase enzyme found in the blood stream that is activated bt HDL apoproteins (apoA-1) adds a fatty acid to cholesterol - this makes soluble cholesteryl esters such as those found in HDL these cholesteryl esters can be transported to other lipoproteins like IDL and then by acquiring those cholesteryl esters they become LDL enzyme that catalyzes cholesterol esterification
answer
LCAT
question
component of all cells plays a major role in the synthesis of cell membranes, steroid hormones, bile salts and vitamin D
answer
cholesterol
question
most cells get their cholesterol from LDL and HDL may be synthesized de novo
answer
sources of cholesterol
question
occurs in the liver driven by acetyl-CoA and ATP *citrate shuttle* carries mitochondria acetyl-CoA into the cytoplasm in the cytoplasm is where acetyl CoA is converted to cholesterol, and NADPH from PPP supplies the reducing equivalents synthesis of *mevalonic acid* in the SER is the rate limiting step which is catalyzed by *3 hydroxyl 3 methylglutaryl (HMG) CoA reductase*
answer
de novo synthesis of cholesterol
question
1. synthesis can inhibit further synthesis by an feedback inhabitation mechanism =}++++ 2. insulin cholesterol synthesis 3. de novo cholesterol is dependent on the regulating of HMG CoA reductase gene expression in the cell
answer
cholesterol synthesis regulation
question
cholesteryl ester transfer protein facilitates the process of transferring cholesteryl esters from HDL to IDL, which then becomes LDL by getting the esters
answer
CETP
question
long chain carboxylic acids carboxyl carbon is carbon 1 carbon 2 = *alpha carbon* these found within the body occur as salts and are capable of forming micelles or they can be esterfied into other compounds (membrane lipids)
answer
fatty acids
question
used for unsaturated fatty acids omega described the position of the last double bond relative to the end of the chain identifies the major precursor fatty acid
answer
omega numbering system
question
more often called palmitate synthesis (palmitate is the only fatty acid that we can synthesize in de novo) excess carbohydrates and protein through the diet can be converted to fatty acids happens in the liver and the products are transferred to adipose tissue for storage major enzymes: acetyl CoA carboxylase fatty acid synthase both are activated by insulin palitic acid/ palmitate is the primary end product acetyl- CoA shuttling acetyl CoA accumulates in the matrix after a meal - needs to be moved to the cytosol for fatty acid synthesis when isocitrate dehydrogenase slows down due to be energetically satisfied - there is an accumulation of citrate Citrate moves across the membrane *citrate lyase* splits citrate into acetyl CoA and oxaloacetate back in the cytosol oxaloacetate goes back to the mitochondria to continue to move acetyl CoA acetyl CoA carboxylase buts fatty acids and acetyl CoA together - activating acetyl coA fatty acid synthase malonly CoA attaches to the ACP - activating it carbonyl group goes to a hydroxyl group and CO2 is lost (NADPH --> NADP+) dehydration - water is lost double bond is reduced and fatty acid is formed (NADPH)
answer
fatty acid synthesis
question
lipid and carbohydrate synthesis process that does not rely directly on the coding of nucleic acids (unlike protein and nucleic acids synthesis)
answer
non-template synthesis
question
major enzyme in fatty acid synthesis activates acetyl CoA in the cytosol by incorporating it with fatty acids requires biotin and ATP adds CO2 to acetyl CoA --> *malonly CoA* activated by insulin and citrate the CO2 is never really incorporated into the fatty acid because it is removed during the addition of the activated acetyl group to the fatty acid
answer
acetyl CoA carboxylase
question
major enzyme complex in fatty acid synthesis found in the cytosol becomes rapidly induced in the liver following a high meal in carbohydrates - because of the elevated insulin levels contains acyl carrier protein (ACP) pantothenic acid - vitamin B5 NADPH - needed to reduce acetyl groups added to the fatty acid malonly CoA attaches to the ACP - activating it carbonyl group goes to a hydroxyl group and CO2 is lost (NADPH --> NADP+) dehydration - water is lost double bond is reduced and fatty acid is formed (NADPH)
answer
fatty acid synthase
question
reverse process of Beta oxidation triglycerides are the storage form of fatty acids formed by attaching three fatty acids (as fatty acetyl CoA) to glycerol glycerol 3 phosphate is the main enzyme occurs mainly in the liver and somewhat in adipose tissue in liver they are packaged and sent to the adipose tissue via VLDL's
answer
triglyceride synthesis
question
reverse process of triglyceride synthesis happens in the mitochondria matrix and sometimes in peroxisomes insulin inhibits this glucagon stimulates this activation: -fatty acid are metabolized they attach to acetyl CoA to become activated. This is catabolized by the fatty-acyl-CoA synthetase -product is fatty acyl CoA or acyl CoA (acetyl CoA (two carbons attached or palmitoyl CoA has 16 carbons attached) entry into the mitochondria: if longer carbon chains they get transported through the carnitine shuttle *carnitine acyltransferase 1* is the RATE LIMITING enzyme - transports the fatty acid on the carnitine so it can be shuttled into the matrix in matrix: releases molecules of acetyl CoA repetition of the four steps: 1. Oxidation of the fatty acid to form a double bond 2. Hydration of the double bond to form a hydroxyl group 3. Oxidation of the hydroxyl group to form a carbonyl/ beta ketoacid 4. Splitting of the Beta ketoacid into the shorter acyl CoA and acetyl CoA -- each cycle of the four steps releases: 1 acetyl CoA --> enters the citric acid cycle (stimulates gluconeogenesis by activating pyruvate carboxylase, can be used to form ketone bodies , or to start the citric acid cycle) reduces NAD+ and FAD creating NADH and FADH2 NADH and FADH2 than can go into the ETC to create ATP
answer
Beta Oxidation
question
break down of branched fatty acids
answer
alpha oxidation
question
happens in the ER break down of fatty acids creates dicarboxylic acids
answer
omega oxidation
question
activates fatty acids by attaching them to acetyl CoA
answer
fatty-acyl-CoA synthetase
question
RATE LIMITING enzyme in Beta oxidation transports the fatty acid on the carnitine so it can be shuttled into the matrix
answer
carnitine acyltransferase 1
question
results in one acetyl CoA and one propinoly CoA instead of two acetyl CoA propinoly CoA --> methylmalonyl CoA by *propinoyl CoA carboxylase* methylmalonyl CoA --> succinyl CoA by *methylmalonyl mutase* succinyl CoA is a TCA cycle intermediate - can be converted to malate to enter the gluconeogenic pathway in the cytosol - thus this is the exception and can be converted to glucose!
answer
Beta oxidation with off number of carbons
question
coverts propinoly CoA to methylmalonyl CoA when a odd number of carbons is in the fatty acid during Beta oxidation requires biotin/ vitamin B7
answer
propinoyl CoA carboxylase
question
biotin
answer
vitamin B7
question
converts methylmalonyl CoA to succinyl CoA when a odd number of carbons is in the fatty acid during Beta oxidation requires cobalamin/ vitamin B12
answer
methylmalonyl mutase
question
cobalamin
answer
vitamin B12
question
since there are double bonds the enzyme can only hold one double bond in the active site one at a time double bond must be at the 2,3 position enzymes are there to move the double bond position enzymes: enoyl-CoA isomerase 2,4 dienoyl CoA reducatse
answer
Oxidation of saturated fatty acids
question
enzyme that rearranges cis double bonds at the 3.4 position to trans double bond at the 2,3 position
answer
enoyl-CoA isomerase
question
enzyme used to convert two conjugated double bonds to one double bond at the 3,4 position - where enoyl CoA isomerase can act upon it
answer
2,4 dienoyl CoA reducatse
question
liver converts excess Actyl- Coa from Beta oxidation of fatty acids to these two different ones: acetoacetate 3-hydroxybutyrate these can be used for energy for other tissues cardiac, skeletal and renal cortex can use these to go back into acetyl CoA - will use them as soon as they are metabolized in the blood stream to prevent a build up in the blood in starvation state they are high enough of concentration in the blood that the brain can metabolize them
answer
ketone bodies
question
occurs in the mitochondria of liver cells happens when excess acetyl coA is present in the fasting state due to Beta oxidation of fatty acids HMG CoA synthase forms HMG CoA from acetyl CoA HMG CoA lyase forms acetoacetate from HMG CoA acetoacetate spontaneously forms into 3-hydroxybutyrate acetone is a major side product of this but will not be used for energy (dragon breath) both acetoacetate and 3 hydroxybutyrate goes into the blood
answer
ketogenesis
question
3 hydroxybutyrate will be converted to acetoacetate (NAD+ --> NADH) NADH can be used to get ATP in the ETC acetoacetate will be activated in the mitochondria matrix to acetoacetyl-CoA by *succinyl-CoA acetoacetyl-CoA transferase*
answer
ketolysis
question
forms HMG CoA in ketogenesis
answer
HMG CoA synthase
question
breaks down HMG CoA into acetoacetate this acetoacetate can be broken down into 3 hydroxybutarate
answer
HMG CoA lyase
question
commonly known as *thiophorase* will convert acetoacetate to acetoacetyl-CoA happens in the mitochondria matrix during ketolysis this enzyme is only present in tissues other than the liver - will not break down ketones in the liver that were just produced in the liver
answer
succinyl-CoA acetoacetyl-CoA transferase
question
when the body has been in ketosis for longer than a week - 2/3 of of the energy comes from ketone bodies when ketones are broken down into acetyl CoA, pyruvate dehydrogenase will be INHIBITED glycolysis and glucose uptake by the brain will be inhibited
answer
ketolysis in the brain
question
starts in the stomach with pepsin pancreatic enzymes: trypsin, chymotrypsin, carboxypeptidases A and B = all released as zymogens digestion is completed with dipeptidase and aminopeptidase (brush boarder enzymes) after digestion you only have amino acids, dipeptideas and tripeptides proteins go through the lumen via *secondary transport with sodium* goes into the blood stream via *facilitated diffusion* proteins are metabolized in the liver or muscle amino acids that break away from the protein will lose their amino group - transamination or deamination - remaining carbon skeleton will be used for energy -- amino acids that go through this pose a potential threat due to deamination causes a toxin in the body as a form of ammonia - leaves through the urea cycle fate of the side chain depends on the chemistry
answer
protein breakdown
question
biological systems are considered this they can exchange both energy and matter with the enviroment energy is exchanged in the form of mechanical work (moved over a distance) OR is in the form of heat matter is exchanged through food consumption and elimination and respiration
answer
open systems
question
when the biochemical systems are studied on a cellular/subcellular level there is no exchange of matter with the enviroment U or internal energy can only come in the form of work or heat thus U = Q - W (first law of thermodynamics) pressure and volume are constant in most living systems (work is the changes in pressure ad volume) since U can only come from work or heat in this case - you only need to focus on heat in living closed systems
answer
closed systems
question
U sum of all the different interactions between and within atoms in a system energies that contribute: vibration rotation linear motion stored chemical
answer
internal energy
question
U = Q - W work refers to the changed in pressure and volume
answer
first law of thermodynamics
question
measures the overall change in heat of a system during a reaction enthalpy at constant pressure and volume, this and Q are equal
answer
????H
question
used to describe energy states in biological systems ATP plays a crucial role in transferring energy from energy releasing catabolic processes to energy requiring anabolic processes whether or not a reaction proceeds is determined by the degree at which enthalpy and entropy change during the chemical reaction
answer
bioenergetics
question
????G changes in this provide information about the chemical reactions and can predict whether a chemical reaction is favorable and will occur this will predict what direction the chemical reaction will go spontaneously *predicts changes occurring at any concentration of products and reactants and at any temperature* a shift in this as a result in concentration is not away or toward spontaneity - = net loss of free energy, spontaneous reaction that will go forward more products than reactants usually will have a +E = electromotive force + = net gain of energy, non-spontaneous reaction that will go backwards more reactants than products usually will have a -E = electromotive force as this goes toward 0 - the reaction proceeded to equilibrium and there s no net change on concentration of reactants and products
answer
change in free energy
question
release energy breaking down
answer
catabolic process
question
need energy building
answer
anabolic process
question
thermodynamic heat exchange equal to enthalpy/ H when pressure and volume are constant
answer
Q - thermodynamics
question
entropy changes in this measure the degree of disorder or energy dispersion in a system chaos
answer
????S
question
represented by ????G°'
answer
physiological conditions
question
Standard Free energy energy change that occurs at standard concentrations of 1M, pressure 1 atm and temperature 25°C ????G = ????G° + RT ln (Q) R= gas constant T = temperature Q = reaction quotient this works for physiological conditions besides the pH/ 1M concentration 1M concentration correlates to a 0 pH - not an physiological pH
answer
????G°
question
modified standard state has everything in standard free energy but has a ph = 7 [H+]= 10^-7 saying that the standard free energy is standardized to the neutral buffers used in biochemistry
answer
????G°'
question
mid level energy carrier formed by substrate level phosphorylation or oxidative phosphorylation ATP cannot get the left over free energy after a reaction - gives about 30 kj/mol.... if reaction only requires 10kj/mol 20 kj/mol is wasted - reason why it is the primary energy carrier and not cAMP who can give -50kj/mol generated from ADP and Pi with energy input from an exergonic reaction or electrochemical gradient ATP is consumed through hydrolysis or a transfer of a phosphate group to another molecule uses about 90% of body weight in ATP - continuos recycling of this provides its energy through the high energy phosphate bonds negative charges on the phosphate groups experience repulsion forces with one another ADP and Pi are stabilized through resonance ATP is much more stable after hydrolysis negative ????G because it wants to break down, and be more stable as it loses its phosphate groups the ????G becomes more positive
answer
ATP as an energy carrier
question
is part of a coupled reaction as an energy source
answer
ATP hydrolysis
question
many of these use ATP as their energy source ex. movement of sodium and potassium against their electrochemical gradient requires energy - harnessed from the hydrolysis of ATO
answer
coupled reactions
question
transfer of a high energy phosphate group from ATP to another molecule this activates or inactivates the target molecule this leads to a phosphoryl group transfer - the overall free energy of the reaction will be determined by taking the sum of the energies of the individual reactions
answer
ATP cleavage
question
the overall free energy of the reaction will be determined by taking the sum of the energies of the individual reactions - due to a transfer of phosphate groups from ATP free energy of hydrolysis (transfer of phosphate group to water) ATP + H2O --> ADP + Pi ????G°'= -30kj/mol
answer
phosphoryl group transfers
question
applies for all of the state functions (pressure, density, temperature, volume, enthalpy, internal energy, free energy, entropy) help to determine the difference in free energy between reactants and products

answer
Hess's Law
question
these components help determine how many electrons have been transferred
answer
Half reactions
question
all soluble NADH, NADPH, FADH2, ubiquinone, cytochromes, glutathione, FMN, iron sulfer clusters
answer
electron carriers
question
contain modified B2/ riboflavin either FAD or FMN most notable for their presence in mitochondria and chloroplasts as electron carriers also involved in modification og other B vitamins to their active forms they function as coenzymes for enzymes in the oxidation of fatty acids, the decarboxylation of pyruvate and the reduction of glutathione
answer
favoproteins
question
vitamin B2
answer
riboflavin
question
physiological tendency toward a relative stable state that is maintained and adjusted often with expenditure of energy equilibrium is a fixed state - which prevents us from storing and energy for later use or creating an excitable enviroment this allows us to store energy for later use
answer
homeostasis
question
absorptive state well fed state occurs shortly after eating marked by greater *anabolism* - synthesis of molecules usually lasts three to five hours after a meal blood glucose releases insulin insulin targets the liver, adipose tissue and muscle insulin promotes glycogen synthesis in the liver and muscle once glycogen stores are filled, the liver converts the excess glucose to fatty acids and triglycerides insulin promotes triglyceride synthesis in adipose tissue and protein synthesis in muscle also allows glucose to enter both tissues liver energy needs are met by oxidation of excess amino acids nervous tissue and RBC are insensitive to insulin nervous tissue gets it energy from oxidizing glucose to CO2 and water RBC use glucose anaerobically for all their energy needs
answer
postprandial state
question
fasting state counterregulatory hormones are released liver glucogen degradation and release of glucose into the blood - glucogenolysis (faster) hepatic gluconeogenesis is stimulated by glucagon - takes about 12 hours to hit maximum velocity release of amino acids from skeletal muscle and fatty acids from adipose tissue both stimulated by the decrease of insulin and increased levels of epinephrine -once the amino acids and fatty acids are carried to the liver they can give the carbon skeleton necessary for gluconeogenesis nervous tissue gets it energy from oxidizing glucose to CO2 and water RBC use glucose anaerobically for all their energy needs
answer
Postabsorptive state
question
glucagon cortisol epinephrine norepinephrine growth hormone have the opposite effects of insulin
answer
counterregulatory hormones
question
starvation glucagon and epinephrine are at their highest results in the rapid degradation of glycogen stores in the liver gluconeogenic activity maintains the blood glucose levels after 24 hours - gluconeogenesis is the primary source of glucose for the body lipidlysis is rapid - resulting in excess acetyl CoA - used to make ketone bodies once fatty acids and ketone bodies are high enough, the muscle will utilize the fatty acids as its major food source and the brain will adapt to using ketones for its energy the shift from glucose to ketones as the major fuel will reduce the quantity of amino acids that must be degraded to support gluconeogenesis RBC will still use glucose anaerobically for all their energy needs
answer
prolonged fating state
question
peptide hormone secreted by the Beta cels of the pancreatic islets of Langerhans key player in the uptake and storage of glucose glucose uptake by these tissues is NOT effected by insulin: nervous tissue kidney tubules intestinal mucosa RBC Beta cells of the pancreas --- these tissues must still be able to absorb glucose even when the glucose concentration is low carbohydrates- insulin increases the uptake of glucose in muscle and can be used as additional fuel in muscle and in fat increased glucose in the muscle can be used as additional fuel to burn in exercise or can be stored as glycogen increases glycogen synthesis in the liver by increases the activity of *glucokinase* and *glycogen synthase* insulin increases glycogen synthesis in the liver by decreasing the activity of glycogen breakdown through *glycogen phosphorlyase* and *glucose-6-phosphatase* proteins increases amino acid uptake by muscle cells increases protein synthesis and decreases the breakdown of essential proteins insulin secretion is directly proportional to blood glucose glucose must enter the Beta islet cells to increase the ATP intracellular concentration increased ATP production leads to calcium release - which then allows the insulin which is encased in vesicles to be exocytosized
answer
insulin
question
1. glucose and triglyceride uptake by fat cells 2. lipoprotein lipase activity - which clears the VLDL and chylomicrons from the liver 3. triglyceride synthesis (lipogenesis) in adipose tissue and the liver from acetyl coA 4. glycogen synthesis by increasing activity of glucokinase and glycogen synthase 5. uptake of amino acids by the muscle cells to increase protein synthesis
answer
insulin increases
question
1. triglyceride breakdown/lipolysis in adipose tissue 2. formation of ketone bodies in the liver
answer
insulin decreases
question
peptide hormone secreted by the alpha cells of the pancreatic islets of langerhans glucagon primarily targets the hepatocyte/ liver cell causes: 1.increased liver glycogenolysis by activating glycogen phosphoroylase and inactivates glycogen synthase 2. increased liver gluconeogenesis by promoting the conversion of pyruvate to phosphoenolpyruvate by *pyruvate carboxylase* and *PEPCK* 2. Also increases liver gluconeogenesis by increasing the conversion of fructose 1,6 bisphosphate to fructose 6 phosphate by *fructose 1,6 biphosphatase* 4. increases lipolysis in the liver by activating *hormone sensitive lipase* - because its effect is on the liver and not the fat cell, it is *NOT considered a major fat mobilizing hormone* when have low blood sugar/hypoglycemia it promotes the secretion of this basic amino acids (arginine, lysine, histidine) can promote the secretion of this - this can be secreted in response to the protein rich meals
answer
glucagon
question
insulin associated with the well fed state glucagon associated with the postabsorptive metabolic state they oppose each other enzymes that are phosphorylated by glucagon are generally dephosphorylated by insulin and vise versa
answer
functional relationship between glucagon and insulin
question
they come from the adrenal cortex in response to stress during flight or fight response - glucose must be rapidly metabolized from the liver to fuel the activity of the contracting muscles while the fatty acids are being released from the adipose tissue example. cortisol secreted in many forms of stress (exercise, cold, emotional stress)
answer
glucocorticoids
question
glucocorticoid steroid hormone promotes the mobilization of energy stores through the degradation and increased delivery of amino acids and increase lipolysis elevates the blood glucose levels increases glucose availability for nervous tissue 1. inhibits glucose uptake in most tissues (muscle, lymphoid, fat) and increases the liver output of glucose through gluconeogenesis 2. enhances the activity of glucagon, epinephrine and other catecholamines prolonged this can causes persistant hyperglycemia which then stimulates insulin and that actually promotes lipogenesis rather than lipolysis
answer
cortisol
question
secreted by the adrenal medulla ex. epinephrine norepinephrine increase the activity of liver and muscle glycogen phosphorylase - promotes glucogenolysis - increasing the rate at which the liver can release glucose into the blood glycogenolysis also increases in the muscle muscle lacks glucose 6 phosphatase which means that glucose cannot be released by skeletal muscle into the blood stream - glucose here is metabolized by the muscle itself act on adipose tissue to increase lipolysis by increasing the activity of hormone sensitive lipase the glycerol and triglyceride breakdown are substrates for the gluconeogenesis
answer
catecholamines
question
type of catecholamine adrenaline acts on the heart to increase the basal metabolic rate through the sympathetic nervous system - increase is associated with the adrenaline rush needs thyroid hormones to have an effect
answer
epinephrine
question
type of catecholamine noradrenaline
answer
norepinephrine
question
these hormones are kept very constant they increase the basal metabolic rate increase O2 consumption and heat production when they are secreted primary effects on carbohydrate and lipid metabolism accelerate cholesterol clearance from the plasma and increase the rate at which glucose is absorbed from the small intestine epinephrine requires these to have an effect
answer
thyroid hormones
question
T4 one of the thyroid hormones increases the metabolic rate at a much slower rate but its effects can last for several days precursor to T3
answer
thyroxine
question
T3 one of the thyroid hormones increases the metabolic rate at a much faster response last over a much shorter duration
answer
triiodothyroxine
question
enzymes that remove iodine from a molecule can make T4 into T3
answer
deiodonases
question
major role is to maintain a constant level of blood glucose under a wide range of conditions and to synthesize ketones when excess fatty acids are being oxidized after a meal... takes glucose and uses it to replenish the glycogen stores remaining glucose is used to form into acetyl CoA and used for fatty acid synthesis fatty acids are converted into triglycerides and released as VLDL well fed state gets most of its energy from the oxidation of amino acids between meals/ fasts releases glucose into the blood increase in glucagon promotes glycogenolysis and gluconeogenesis lactate from anaerobic metabolism, glycerol from triglycerides and amino acids provide the carbon skeleton for glucose synthesis
answer
liver
question
after a meal insulin stimulate glucose uptake by this tissue insulin also triggers fatty acid released from VLDL and chylomicrons - which carry the triglycerides absorbed from the gut lipoprotein lipase is also induced by insulin fatty acids that are released from the lipoproteins are taken up by the adipose tissue and re-esterfied to triglycerides for storage *glycerol phosphate* which is required for the triglyceride synthesis comes from glucose that is metabolized in adipocytes as an alternative product of glycolysis prolonged fast decreased levels of insulin, increased levels of epinephrine activate the hormone sensitive lipase in fat cells - allows the fatty acids to be released into circulation
answer
adipose tissue
question
enzyme found in the capillary beds of adipose tissue induced by insulin
answer
lipoprotein lipase
question
major fuels for this are glucose and fatty acids due to the size of muscle - it needs the most fuel from the body insulin promotes glucose uptake by it to replenish glycogen stores and amino acid synthesis excess glucose and amino acids can be oxidized for energy fasting state uses the fatty acids from the blood stream as energy can also use ketones for energy
answer
resting muscle
question
short burst/high intensity energy demands -creatine phosphate gives the 2-7 short bust of energy (transferring a phosphate to ADP to create ATP) the muscle stores both glycogen and triglycerides -blood glucose and free fatty acids may be used for energy -anaerobic glycolysis drawing from stored glycogen high intensity continuos energy demands oxidation of glucose and fatty acids after 1-3 hours glycogen stores are depleted and the exercise declines to the rate that can be supported by oxidation of fatty acids
answer
active muscle
question
fatty acids are their major fuel source!!! - even in a well fed state ketones can also be used during prolonged fasting in cardiac hypertrophy (thickening of heart) or failing heart - glucose oxidation increases and Beta oxidation falls
answer
cardiac muscle
question
uses 25% of the total bodies glucose uses 20% of the bodies O2 when hypoglycemic - glucagon and epinephrine is released free fatty acids cannot cross the blood brain barrier - *fatty acids will never be used as an energy source for the brain* glucose is supplied to the brain via gluconeogenesis or glucogenolysis in prolonged fasting ketone bodies can be used 1/3 of the brain will still remain to use glucose though - even in a prolonged fasting state
answer
brain
question
under 70 mg/dL
answer
hypoglycemia
question
accurate measurement of the respiratory quotient
answer
respirometry
question
RQ differs depending on the fuels being used by the organism for carbohydrates it is about 1 for lipids it is about .7 resting people about .8 (both fat and glucose are being consumed) changes under high stress, starvation and different hormones
answer
respiratory quotient
question
measure the BMR based on heat exchange with the enviroment
answer
calorimeters
question
basal metabolic rate can be based on age, weight, height and gender
answer
BMR
question
lipids is the primary factor in the gradual change of body mass over time maintaing weight - eating the same amount of energy that is spent on the average day if body weight increases so does the BMR thus the caloric excess will cause an increase in body mass until equilibrium is reached between the new basal rate and the existing intake body does have a threshold
answer
regulation of body mass
question
small increase/decrease in activity - compensations in hunger, won't be as hungry or will be more hungry alterations to body mass happen is the alterations are above the threshold
answer
hunger threshold
question
now hungry due to ghrelin and orexin
answer
not hungry - walk into favorite restaurant
question
secreted by the stomach in response to a signal that you will soon be eating sight, sound, taste and smell act as signals to release this increases appetite and stimulates the secretion of orexin
answer
ghrelin
question
further increases appetite released in response to ghrelin involved in the alertness and sleep wake cycle hypoglycemia also triggers this to be released
answer
orexin
question
hormone secreted by fat cells decreases appetite suppresses orexin production mutations in this can lead to obesity
answer
leptin
question
body mass index normal 18-25 25-30 overweight 30 obese
answer
BMI
question
help the substrate better fit to the enzyme small organic groups vitamins NADH coenzyme Q help substrate better fit the enzyme can bind to the active site of the enzyme and participate in the catalysis of the reaction usually *carry charge* through ionization, protonation, deprotonation usually kept at low concentrations - recruited only when needed can be attached via weak covalent bonds or by strong covalent bonds
answer
coenzymes
question
C and B need to be replenished coenzyme
answer
water soluble vitamins
question
DEAK better regulated by partition coefficients coenzyme fat frat football guys
answer
fat soluble vitamins
question
complementary DNA formed in the processed mRNA strand by reverse transcription used in DNA libraries contains only the exons which where transcriptionally active
answer
cDNA



