DL-AP4 C4H10NO5P structure – Flashcards
Flashcard maker : Henry Smith
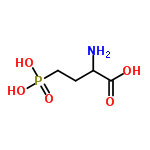
| Molecular Formula | C4H10NO5P |
| Average mass | 183.100 Da |
| Density | 1.6±0.1 g/cm3 |
| Boiling Point | 491.7±55.0 °C at 760 mmHg |
| Flash Point | 251.2±31.5 °C |
| Molar Refractivity | 35.6±0.3 cm3 |
| Polarizability | 14.1±0.5 10-24cm3 |
| Surface Tension | 88.8±3.0 dyne/cm |
| Molar Volume | 112.4±3.0 cm3 |
- Experimental data
- Predicted – ACD/Labs
- Predicted – EPISuite
- Predicted – ChemAxon
- Experimental Physico-chemical Properties
- Miscellaneous
Predicted data is generated using the ACD/Labs Percepta Platform – PhysChem Module
| Density: | 1.6±0.1 g/cm3 |
| Boiling Point: | 491.7±55.0 °C at 760 mmHg |
| Vapour Pressure: | 0.0±2.7 mmHg at 25°C |
| Enthalpy of Vaporization: | 83.1±6.0 kJ/mol |
| Flash Point: | 251.2±31.5 °C |
| Index of Refraction: | 1.546 |
| Molar Refractivity: | 35.6±0.3 cm3 |
| #H bond acceptors: | 6 |
| #H bond donors: | 5 |
| #Freely Rotating Bonds: | 4 |
| #Rule of 5 Violations: | 1 |
| ACD/LogP: | -2.39 |
| ACD/LogD (pH 5.5): | -6.06 |
| ACD/BCF (pH 5.5): | 1.00 |
| ACD/KOC (pH 5.5): | 1.00 |
| ACD/LogD (pH 7.4): | -6.19 |
| ACD/BCF (pH 7.4): | 1.00 |
| ACD/KOC (pH 7.4): | 1.00 |
| Polar Surface Area: | 131 Å2 |
| Polarizability: | 14.1±0.5 10-24cm3 |
| Surface Tension: | 88.8±3.0 dyne/cm |
| Molar Volume: | 112.4±3.0 cm3 |
Predicted data is generated using the US Environmental Protection Agency’s EPISuite™
Log Octanol-Water Partition Coef (SRC): Log Kow (KOWWIN v1.67 estimate) = -4.03 Boiling Pt, Melting Pt, Vapor Pressure Estimations (MPBPWIN v1.42): Boiling Pt (deg C): 431.18 (Adapted Stein & Brown method) Melting Pt (deg C): 226.38 (Mean or Weighted MP) VP(mm Hg,25 deg C): 5.96E-011 (Modified Grain method) Subcooled liquid VP: 8.61E-009 mm Hg (25 deg C, Mod-Grain method) Water Solubility Estimate from Log Kow (WSKOW v1.41): Water Solubility at 25 deg C (mg/L): 1e+006 log Kow used: -4.03 (estimated) no-melting pt equation used Water Sol Estimate from Fragments: Wat Sol (v1.01 est) = 1e+006 mg/L ECOSAR Class Program (ECOSAR v0.99h): Class(es) found: Aliphatic Amines-acid Henrys Law Constant (25 deg C) [HENRYWIN v3.10]: Bond Method : 3.28E-019 atm-m3/mole Group Method: Incomplete Henrys LC [VP/WSol estimate using EPI values]: 1.436E-017 atm-m3/mole Log Octanol-Air Partition Coefficient (25 deg C) [KOAWIN v1.10]: Log Kow used: -4.03 (KowWin est) Log Kaw used: -16.873 (HenryWin est) Log Koa (KOAWIN v1.10 estimate): 12.843 Log Koa (experimental database): None Probability of Rapid Biodegradation (BIOWIN v4.10): Biowin1 (Linear Model) : 0.8869 Biowin2 (Non-Linear Model) : 0.8968 Expert Survey Biodegradation Results: Biowin3 (Ultimate Survey Model): 3.1836 (weeks ) Biowin4 (Primary Survey Model) : 4.0124 (days ) MITI Biodegradation Probability: Biowin5 (MITI Linear Model) : 0.4300 Biowin6 (MITI Non-Linear Model): 0.2403 Anaerobic Biodegradation Probability: Biowin7 (Anaerobic Linear Model): 1.0863 Ready Biodegradability Prediction: NO Hydrocarbon Biodegradation (BioHCwin v1.01): Structure incompatible with current estimation method! Sorption to aerosols (25 Dec C)[AEROWIN v1.00]: Vapor pressure (liquid/subcooled): 1.15E-006 Pa (8.61E-009 mm Hg) Log Koa (Koawin est ): 12.843 Kp (particle/gas partition coef. (m3/ug)): Mackay model : 2.61 Octanol/air (Koa) model: 1.71 Fraction sorbed to airborne particulates (phi): Junge-Pankow model : 0.99 Mackay model : 0.995 Octanol/air (Koa) model: 0.993 Atmospheric Oxidation (25 deg C) [AopWin v1.92]: Hydroxyl Radicals Reaction: OVERALL OH Rate Constant = 41.0056 E-12 cm3/molecule-sec Half-Life = 0.261 Days (12-hr day; 1.5E6 OH/cm3) Half-Life = 3.130 Hrs Ozone Reaction: No Ozone Reaction Estimation Fraction sorbed to airborne particulates (phi): 0.992 (Junge,Mackay) Note: the sorbed fraction may be resistant to atmospheric oxidation Soil Adsorption Coefficient (PCKOCWIN v1.66): Koc : 31.06 Log Koc: 1.492 Aqueous Base/Acid-Catalyzed Hydrolysis (25 deg C) [HYDROWIN v1.67]: Rate constants can NOT be estimated for this structure! Bioaccumulation Estimates from Log Kow (BCFWIN v2.17): Log BCF from regression-based method = 0.500 (BCF = 3.162) log Kow used: -4.03 (estimated) Volatilization from Water: Henry LC: 3.28E-019 atm-m3/mole (estimated by Bond SAR Method) Half-Life from Model River: 2.415E+015 hours (1.006E+014 days) Half-Life from Model Lake : 2.635E+016 hours (1.098E+015 days) Removal In Wastewater Treatment: Total removal: 1.85 percent Total biodegradation: 0.09 percent Total sludge adsorption: 1.75 percent Total to Air: 0.00 percent (using 10000 hr Bio P,A,S) Level III Fugacity Model: Mass Amount Half-Life Emissions (percent) (hr) (kg/hr) Air 1.2e-009 6.26 1000 Water 39 360 1000 Soil 60.9 720 1000 Sediment 0.0713 3.24e+003 0 Persistence Time: 579 hr
Click to predict properties on the Chemicalize site


